To date, many molecular mechanisms of fluconazole (FLZ) resistance in Candida species have been described and may occur simultaneously in resistant isolates.4 One of the most studied mechanisms is characterized by missense mutations in ERG11 gene, leading to a decrease in the affinity of the FLZ for its enzyme target, 14α-demethylase. The identification of these mutations is based on the sequencing of the ERG11 open reading frame (ORF) followed by computational analysis of translated codons. However, the cost of the sequencing for a large number of samples is high and expensive for many laboratories. The single-strand conformational polymorphism (SSCP) is a mutation-screening technique widely used for its simplicity and high resolution, detecting changes even at a single base level. This ability is because a single-stranded DNA folds differently from another if it differs by a single base, resulting in different mobility in electrophoresis gels.2 In this light, we evaluated the usefulness of SSCP as a screening method to detect mutations in ERG11 gene.
In this study, a total of 87 Candida clinical isolates (32 Candida albicans, 34 Candida tropicalis, 11 Candida parapsilosis complex, six Candida glabrata and four Candida krusei) were evaluated. Molecular identification and susceptibility testing were previously performed.1 To improve the SSCP efficiency, we amplified the ERG11 gene of each species in four PCR products around 400bp using previously described primers.1 These four PCR products cover together the entire ORF. Once amplified, each PCR product was denatured by 15min at 95°C after addition of 1:1 running buffer (formamide 98%, EDTA 10mM, bromophenol blue 0.05% and xylene cyanol 0.05%), chilled by 5min in ice and then loaded in SSCP gels. The electrophoresis were performed in 6% polyacrylamide gels (glycerol 5% added) under the following conditions: 45mA at 10°C by 5.5h in 1× TBE buffer (tris–borate–EDTA). Gels were fixed in ethanol 10% for 30min and then submitted to silver staining.3 The same PCR products were further purified, and both strands sequenced and compared to their respective migration patterns on SSCP gels.
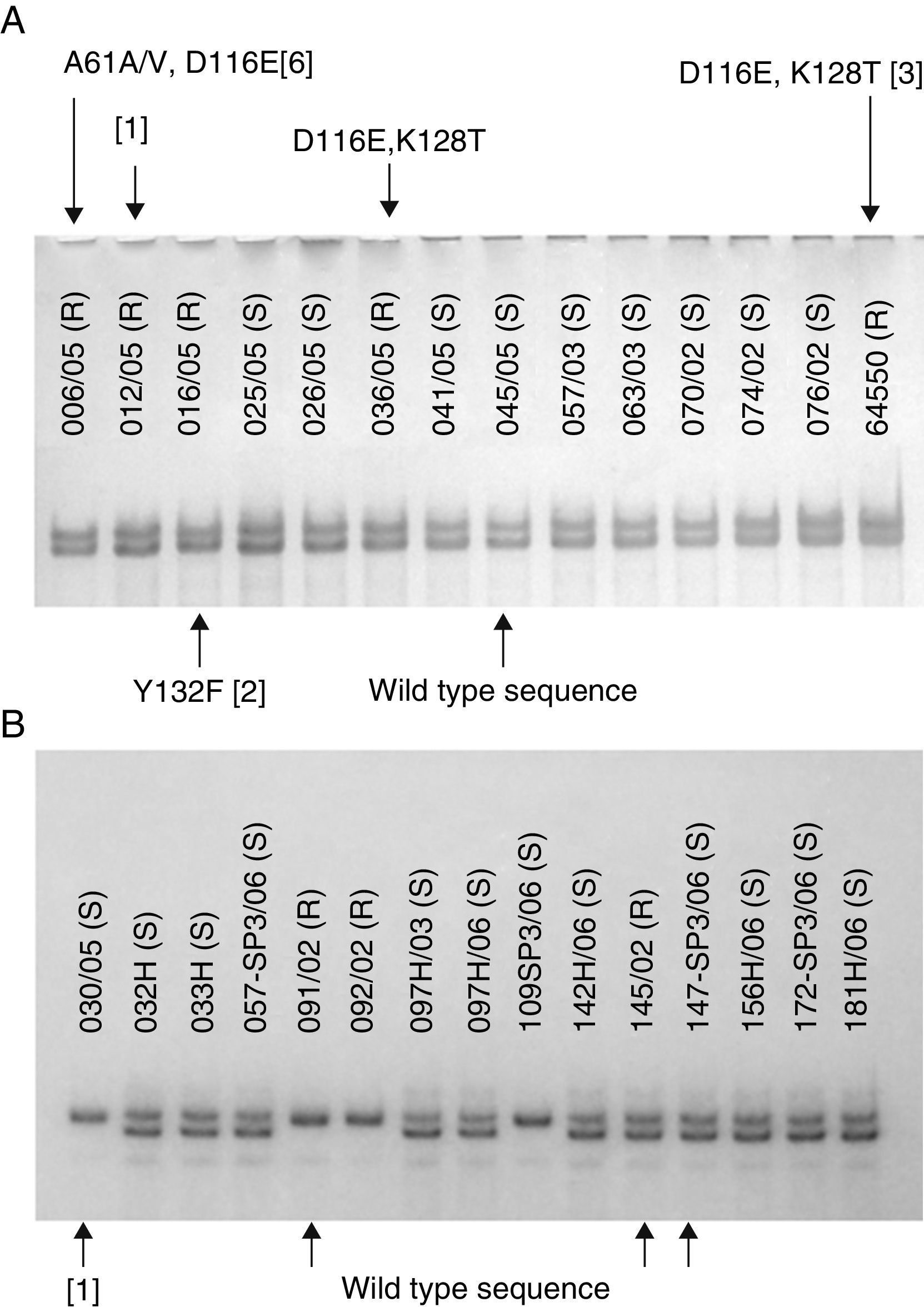
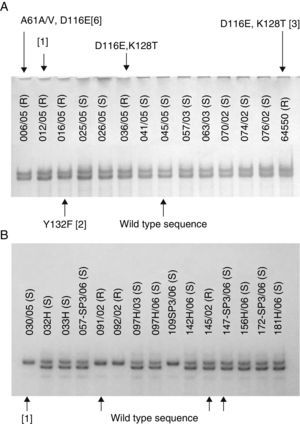
Because the SSCP is able to detect mutations in a single base (point mutations), we expected the isolates with mutated sequences to show different SSCP patterns when compared to the wild sequence isolates. However, the results observed were not the expected, and we could not draw a correlation between SSCP patterns and the presence of mutations. Indeed, we observed that some PCR products with the same SSCP pattern exhibited different number of mutations or even their absence (Fig. 1). In the same way, some different SSCP patterns showed the same wild type sequence. The susceptibility data could not be either linked to SSCP patterns. In other words, susceptible and resistant isolates shared the same patterns. These results were observed in all the four PCR products from all five species, suggesting the SSCP is not a reliable screening technique to detect mutations in ERG11 gene from Candida clinical isolates. On the other hand, different results might be reached when analyzing shorter PCR products (<400bp) once the SSCP sensitivity is highly influenced by both length of the DNA amplicon as well as the GC content.
(A) SSCP gel of the first ∼400bp of ERG11 gene of Candida albicans clinical isolates showing different mutations (indicated by arrows) at the same electrophoresis pattern. (B) SSCP gel of the last ∼400bp of ERG11 gene of Candida tropicalis clinical isolates showing different electrophoresis patterns with the same wild type sequence. Silence mutations are presented in brackets and heterozygosity by a slash.