There is an increasing interest in the study of microorganisms that inhabit extreme environments for reasons that vary from gaining insight into the origin of life to the searching of new biotechnological applications.
AimsIn this work, we studied the tolerance of fungi isolated from the Aguas Agrias Stream (AAS; Tharsis, Huelva, Spain), an acidic metal-rich environment, to a culture medium prepared with water from this extreme ecosystem (AASW medium). The ability of some culture collection strains of moulds and yeasts to grow on AASW medium was also assessed.
MethodsFor moulds, a tolerance index was calculated by dividing the growth diameter of colonies on AASW medium by the diameter in the control medium, and their germinative potential was recorded. For yeasts and yeast-like fungi, the minimum inhibitory concentration of AASW was determined.
ResultsIn general, the fungi isolated from the AAS showed differences in their ability to germinate and grow on AASW medium. Collection strains of the genus Aspergillus could grow on AASW medium, but showed some differences in tolerance when compared to environmental isolates.
ConclusionsExtremotolerant fungi can manifest differences in their tolerance to culture media that simulate the conditions of their natural habitat. The results of this work suggest that the ability of fungi to grow in acidic, metal-rich environments might be more widespread than previously thought, and highlight the importance of determining the factors that are responsible for tolerance to these extreme environments.
Hay un creciente interés por el estudio de los microorganismos que habitan ambientes extremos por razones que van desde incrementar el conocimiento sobre el origen de la vida hasta la búsqueda de nuevas aplicaciones biotecnológicas.
ObjetivosEn el presente trabajo se aborda el estudio de la tolerancia de hongos aislados del Arroyo de Aguas Agrias (AAS; Tharsis, Huelva, España), un ambiente ácido y rico en metales, frente a medios de cultivo preparados con agua procedente de este ecosistema extremo (medio AASW). También se investigó la posibilidad de crecimiento en estas condiciones de cepas de colección de hongos y levaduras.
MétodosPara los hongos miceliares se calculó un índice de tolerancia, definido como el cociente entre el diámetro de crecimiento de las colonias sobre AASW y el que se produce en un medio control. Para las levaduras se determinó la concentración mínima inhibitoria de AASW.
ResultadosEn general, los hongos aislados del AAS manifestaron diferencias en su capacidad para germinar y crecer sobre el medio AASW. Las cepas de colección del género Aspergillus fueron capaces de crecer sobre el medio AASW, pero mostraron diferencias en su tolerancia al mismo en comparación con los aislamientos ambientales.
ConclusionesLos hongos extremotolerantes pueden manifestar diferencias en su tolerancia a medios de cultivo que simulan las condiciones de su hábitat natural. Los resultados de nuestro trabajo sugieren que la capacidad de los hongos para crecer en ambientes ácidos, ricos en metales, puede ser más común de lo que pudiera pensarse, y pone de manifiesto la importancia de determinar los factores específicos que son responsables de la tolerancia a esos ambientes extremos.
There is a growing interest in the study of microorganisms that inhabit extreme environments. This attention stems from the knowledge that studying the microbial ecology of extreme environments provides the limits of life and its possible origin, and because identifying new extremophilic and extremotolerant species and biomolecules is potentially useful for biotechnology.12,24,27,29 Furthermore, the study of extreme ecosystems provides a model for exobiology, as it helps to identify possible habitats for life on other planets.25,29
The Iberian Pyrite Belt (IPB), located in the southwest of the Iberian Peninsula, is rich in complex polymetallic sulfides and is one of the world's most important pyrite regions.10,12,19 Due to the occurrence of acid rock drainage, the main features of the IPB habitats are the low pH and the high concentration of heavy metals.10 The microbiological investigations of the different water courses that flow across the IPB have mainly focused on the spatial and temporal patterns of microbial diversity, and their relationship to different environmental stresses.1–3,5,10,13,22,24 In contrast, the study of the relative tolerance of the different fungal species that inhabit these ecosystems to their environmental conditions has received less attention.
In this work, we studied the tolerance of fungi isolated from the Aguas Agrias Stream (AAS), a mining area located within the IPB, to acidic, metal-rich media prepared with water from this extreme environment (AASW). The ability of some culture collection strains of moulds and yeasts that had never been in contact with the AAS to grow on media containing AASW was also assessed.
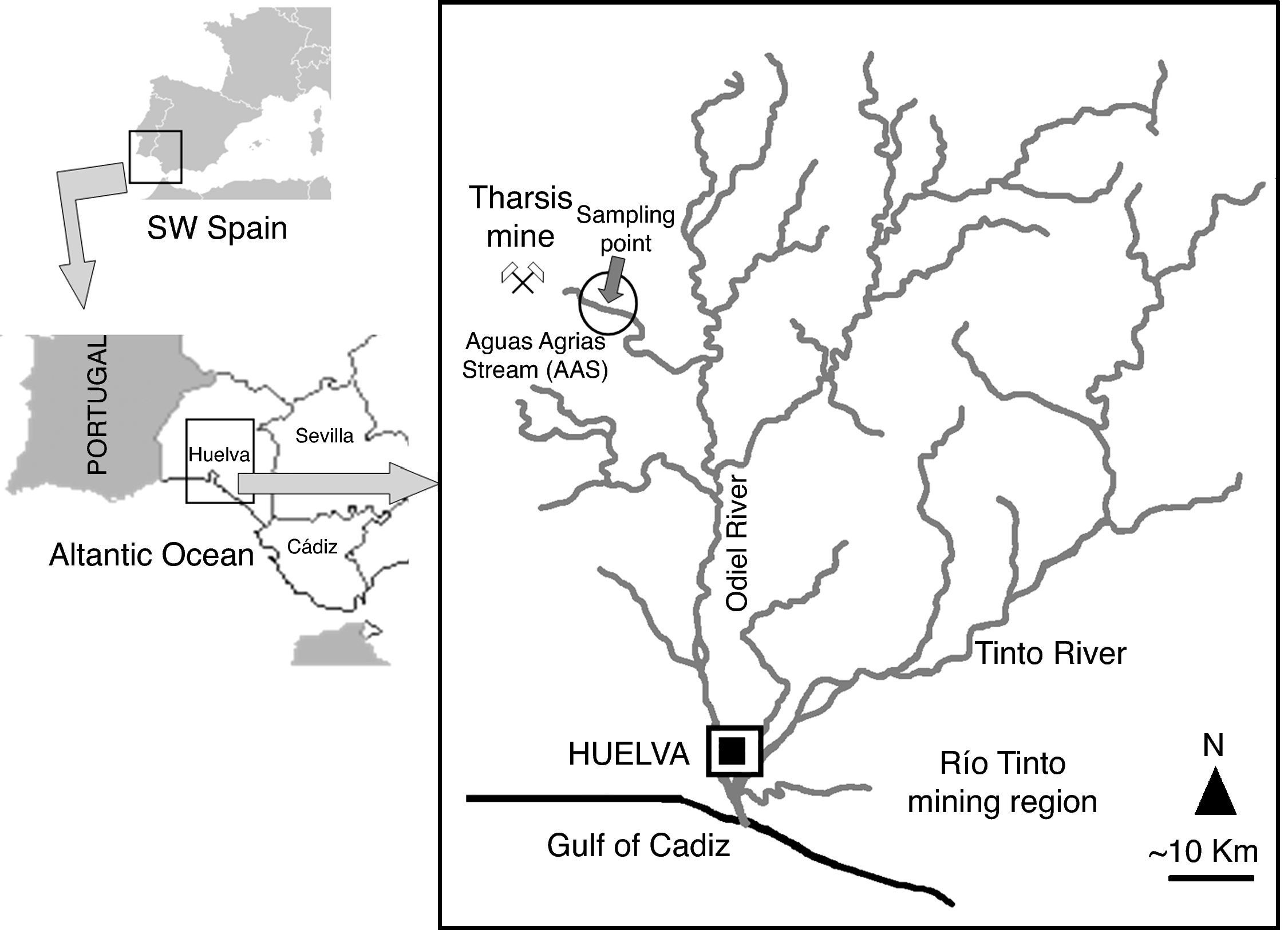
Materials and methodsStudy site and sample collectionSampling was carried out in May 2006, near the Tharsis mine (Huelva, SW Spain; 37°35′45″ N, 07°03′53″ W), an area that has been exploited since ancient times, and over the last two centuries until the 1990s. The approximate geographic location of the Tharsis mine and the sampling point is shown in Figure 1.
Water from the AAS is strongly affected by acid lixiviates from the Tharsis mine, which create a low pH (c. 2.3) and high concentrations of metals (Fe at 1,435mg l-1, Al at 551mg l-1, Zn at 324mg l-1, Mn at 143mg l-1, Cu at 64mg l-1, As (V) at 372μg l-1 and Pb at 650μg l-1).33 Most of the biomass at the AAS is located in the streambed, forming green, dense and compact biofilms.
For microbial analysis, three 50-ml water samples were collected close to the streambanks at 5cm depth, and three 1-cm2 pieces of biofilm were taken from the streambed. All samples were maintained in sterile plastic containers and refrigerated (4°C) until processing, which took place within 24h.
Three AASW samples of 5 l were also collected for selective medium preparation (see below). These samples were homogeneously mixed, and the resulting integrated water sample was filtered (0.22μm, Stericup, Millipore Co., Billerica, MA, USA) and stored in closed bottles at 4°C in the dark until use.
Isolation and identification of moulds and yeastsOne hundred microliters were taken from each water sample and spread on Sabouraud agar plates (bioMérieux, Marcy l’Etoile, France). In addition, a loop of biofilm sample was streaked on Sabouraud agar plates. All plates were incubated at 30°C and observed daily at least for 10 days.
The yeast and mould colonies that appeared on the plates were isolated in pure culture and subcultured on Sabouraud agar, Czapek-Dox Agar (Panreac, Barcelona, Spain) and Potato Dextrose Agar (Pronadisa, Madrid, Spain). Fungi were identified on the basis of macroscopic and microscopic criteria.14,21,28,32,35 All isolates were stored in 10% glycerol at –80°C. To minimize the adaptation of isolates to culture conditions, these frozen stocks were used when necessary to obtain new subcultures on non-selective media.
Genomic DNA isolationA modification of the methodology described by Liu et al.20 was used to obtain DNA from fungal isolates recovered from the AAS. Briefly, a loop of fungal culture was added to microcentrifuge tubes containing 500μl of lysis buffer (400 mmol l-1 Tris-HCl [pH 8.0], 60 mmol l-1 EDTA [pH 8.0], and 150 mmol l-1 NaCl, 1% SDS w/v). The tubes were vortexed briefly and incubated for 2h at 65°C. After adding 150μl of potassium acetate solution (6ml of 5mol l-1 potassium acetate, 1.15ml of glacial acetic acid and 2.85ml of distilled water; pH 4.8), the tubes were vortexed again and centrifuged at 8,000g for 5min. Supernatants were transferred to new tubes and centrifuged as described above. Each supernatant was transferred to another microcentrifuge tube and an equal volume of 2-isopropanol (Sigma-Aldrich, Madrid, Spain) was added. The tubes were mixed by inversion and centrifuged at 16,000g for 15min to precipitate the DNA and discard the supernatant. DNA pellets were washed with 300μl of 70% cold ethanol and centrifuged at 16,000g for 10min and supernatant removed. DNA precipitates were dried at 37°C to remove all traces of ethanol and resuspended in 150μl of sterilized distilled water. DNA concentration and purity were determined by UV spectrophotometry.
Large subunit ribosomal RNA (LSU rRNA) gene sequence analysisThe D1/D2 domains of the large subunit (LSU) rRNA gene were amplified by polymerase chain reaction (PCR) from fungi isolated from the AAS. Reaction mixtures contained genomic DNA (40 to 60 ng), 10 mmol l-1 Tris-HCl (pH 8.3), 50 mmol l-1 KCl, 1.5 mmol l-1 MgCl2, 200μmol l-1 each dNTP, 1.5 U AmpliTaq DNA polymerase (Applied Biosystems, Madrid, Spain) and 20pmol each primer: NL-1 (5′-GCATATCAATAAGCGGAGGAAAAG-3′) and NL-4 (5′-GGTCCGTGTTTCAAGACGG-3′) (Isogen Life Science, Maarssen, Netherlands).17,18 The final volume was adjusted to 50μl. Amplifications were carried out in a GeneAmp PCR System 9700 thermocycler (Applied Biosystems, Foster City, CA, USA) for 5min at 94°C, followed by 35 cycles of 30s at 94°C, 30s at 51°C and 1min at 72°C, and a final extension at 72°C for 10min. The PCR products were purified using the QIAquick PCR purification kit (Qiagen Iberia, Madrid, Spain) according to the manufacturer's protocol. Purified amplicons were sequenced in the forward and reverse directions using the ABI Prism Big Dye Terminator v3.0 Ready Reaction Cycle Sequencing Kit (Applied Biosystems), and analyzed on an ABI Prism 3730 sequencer (Applied Biosystems). The sequences of the D1/D2 domains of the LSU rRNA gene (596-620bp) were compared with those stored in Genbank databases using BLAST software (http://www.ncbi.nlm.nih.gov/blast).
Preparation of selective culture mediaTo study the response of fungi isolated in this work and different culture collection strains to the conditions of the AAS, two kinds of selective culture media were prepared:
- -
Acidic Czapek Agar (ACzA), which is Czapek-Dox Agar (CzA, Pronadisa) adjusted to a final pH of 2.4 with 4 mmol l-1 HCl.
- -
AASW medium, which is CzA containing from 10 to 90% (v/v) AASW. The medium with the highest amount of AASW (90% AASW medium) had a pH of approximately 2.4.
In both cases, to prevent hydrolysis of the medium, the selective agent (HCl or AASW) was filter sterilized and added to the corresponding amount of CzA dissolved in distilled water and sterilized by autoclaving. The media were poured into sterile 90mm Petri dishes and allowed to solidify.
Germination and/or growth ability on ACzA and AASW mediaMouldsThe ability of the moulds isolated from the AAS to germinate and grow on ACzA and 90% AASW media was assessed. Two Aspergillus strains from our culture collection, Aspergillus fumigatus B57098 and Aspergillus niger C2089, were also studied. In all cases, spore suspensions from 10-day plate cultures on Sabouraud agar were prepared in Phosphate Buffered Saline-Tween 20 (0.1%) (PBS-T). Approximately 106 spores from each strain were spread on the surface of the selective media. All the plates were wrapped in aluminium foil to prevent dehydration and incubated at 30°C for at least 10 days.
Yeasts and yeast-like fungiThe ability of environmental isolates of yeasts and yeast-like fungi to form visible colonies on ACzA and 90% AASW medium was assayed. Two yeast strains from our culture collection were also included in the experiment: Rhodotorula mucilaginosa A56A and Cryptococcus laurentii C1077. In all cases, cell suspensions from 72-h plate cultures on Sabouraud agar were prepared in PBS-T. Approximately 107 cells from each isolate were spread on the surface of the selective media and incubation was performed as with moulds.
Tolerance to ACzA and AASW mediaMouldsTolerance to the selective culture media described above was measured using a modification of the procedure described by Valix and Loon.34 A tolerance index was calculated by dividing the mycelial growth diameter on selective medium (ACzA or AASW medium) with the growth diameter on control medium (CzA). To determine this index, 5-mm diameter mycelial disks were taken from the leading edge of 10-day plate cultures of each isolate on Sabouraud agar. Disks were transferred under sterile conditions onto selective media, and all the plates were wrapped in aluminium foil to prevent dehydration and incubated at 30°C for at least 30 days. Mycelial growth diameters were measured in two perpendicular directions, and the mean value was calculated.
Yeasts and yeast-like fungiFor some selected yeast and yeast-like isolates from the AAS or our culture collection (see Table 1 of the Results section), the minimum inhibitory concentration of AASW, which was defined as the minimum concentration of AASW that inhibited the formation of visible colonies, was determined. Cell suspensions from 72-h plate cultures on Sabouraud agar were prepared in PBS-T, and approximately 107 cells were spread on the surface of culture media prepared with AASW at 0 (control medium), 10, 20, 30, 40, 50, 60, 70, 80 or 90% (v/v). Inoculated plates were wrapped in aluminium foil to prevent dehydration and incubated at 30°C for at least 14 days.
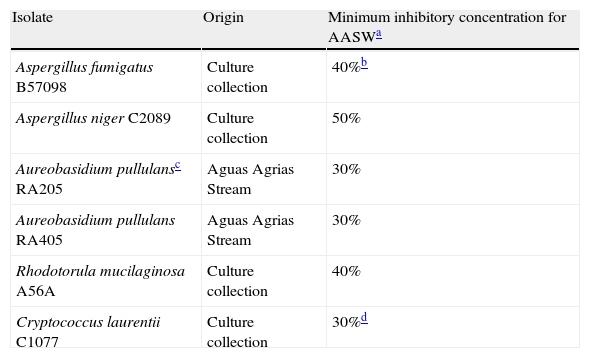
Susceptibility of some selected environmental isolates and culture collection strains to Aguas Agrias Stream water.
| Isolate | Origin | Minimum inhibitory concentration for AASWa |
| Aspergillus fumigatus B57098 | Culture collection | 40%b |
| Aspergillus niger C2089 | Culture collection | 50% |
| Aureobasidium pullulansc RA205 | Aguas Agrias Stream | 30% |
| Aureobasidium pullulans RA405 | Aguas Agrias Stream | 30% |
| Rhodotorula mucilaginosa A56A | Culture collection | 40% |
| Cryptococcus laurentii C1077 | Culture collection | 30%d |
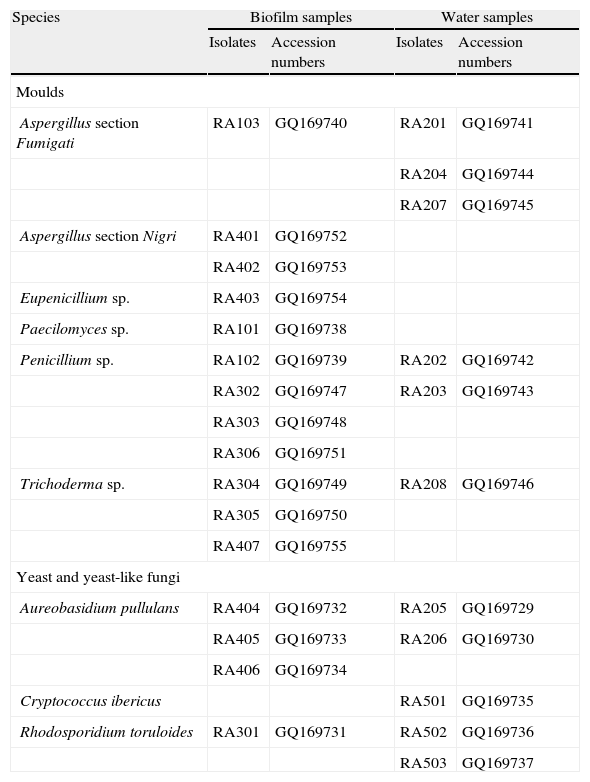
The fungi isolated from the AAS are presented in Table 2. A total of 18 mould isolates were recovered. Only one was a teleomorph, and belonged to the ascomycetous genus Eupenicillium. In most cases, the macroscopic and microscopic characterization of the isolates allowed the identification to the genus level only. Furthermore, molecular identification by sequencing of the D1/D2 domains of the LSU rRNA gene was also inconclusive at the species level, because of the high similarity (≥ 99% identity) between the query sequences and sequences deposited in the GenBank that belonged to different species. The accession numbers of the fungal isolates characterized in this work are shown in Table 2.
Fungi isolated from the Aguas Agrias Stream.
| Species | Biofilm samples | Water samples | ||
| Isolates | Accession numbers | Isolates | Accession numbers | |
| Moulds | ||||
| Aspergillus section Fumigati | RA103 | GQ169740 | RA201 | GQ169741 |
| RA204 | GQ169744 | |||
| RA207 | GQ169745 | |||
| Aspergillus section Nigri | RA401 | GQ169752 | ||
| RA402 | GQ169753 | |||
| Eupenicillium sp. | RA403 | GQ169754 | ||
| Paecilomyces sp. | RA101 | GQ169738 | ||
| Penicillium sp. | RA102 | GQ169739 | RA202 | GQ169742 |
| RA302 | GQ169747 | RA203 | GQ169743 | |
| RA303 | GQ169748 | |||
| RA306 | GQ169751 | |||
| Trichoderma sp. | RA304 | GQ169749 | RA208 | GQ169746 |
| RA305 | GQ169750 | |||
| RA407 | GQ169755 | |||
| Yeast and yeast-like fungi | ||||
| Aureobasidium pullulans | RA404 | GQ169732 | RA205 | GQ169729 |
| RA405 | GQ169733 | RA206 | GQ169730 | |
| RA406 | GQ169734 | |||
| Cryptococcus ibericus | RA501 | GQ169735 | ||
| Rhodosporidium toruloides | RA301 | GQ169731 | RA502 | GQ169736 |
| RA503 | GQ169737 | |||
Four yeast isolates and five isolates of the yeast-like fungus Aureobasidium pullulans were also recovered from the AAS (see Table 2). Analysis of the sequence of the D1/D2 domains of the LSU rRNA gene revealed that three AAS yeast isolates belonged to the species Rhodosporidium toruloides. The fourth isolate, recovered from a sample of water but not from biofilm, showed a 100% DNA sequence similarity with the type strain of Cryptococcus ibericus.
In all cases, fungal growth was accompanied by the growth of a considerable diversity of bacterial types (mainly actinobacteria and Gram-negative and Gram-positive bacilli).
All the mould isolates used in this work, whether isolated from the AAS or from our collection, were capable of germinating and growing on both control and ACzA media. However, only four isolates from biofilm samples, two isolates of Penicillium sp. (RA303 and RA306) and two belonging to the section Nigri of the genus Aspergillus (RA401 and RA402), germinated and formed colonies on 90% AASW medium after a 10-day incubation period. None of the 16 remaining isolates (14 from the AAS and two from our collection) were able to germinate on this medium, even when incubation was extended up to 30 days.
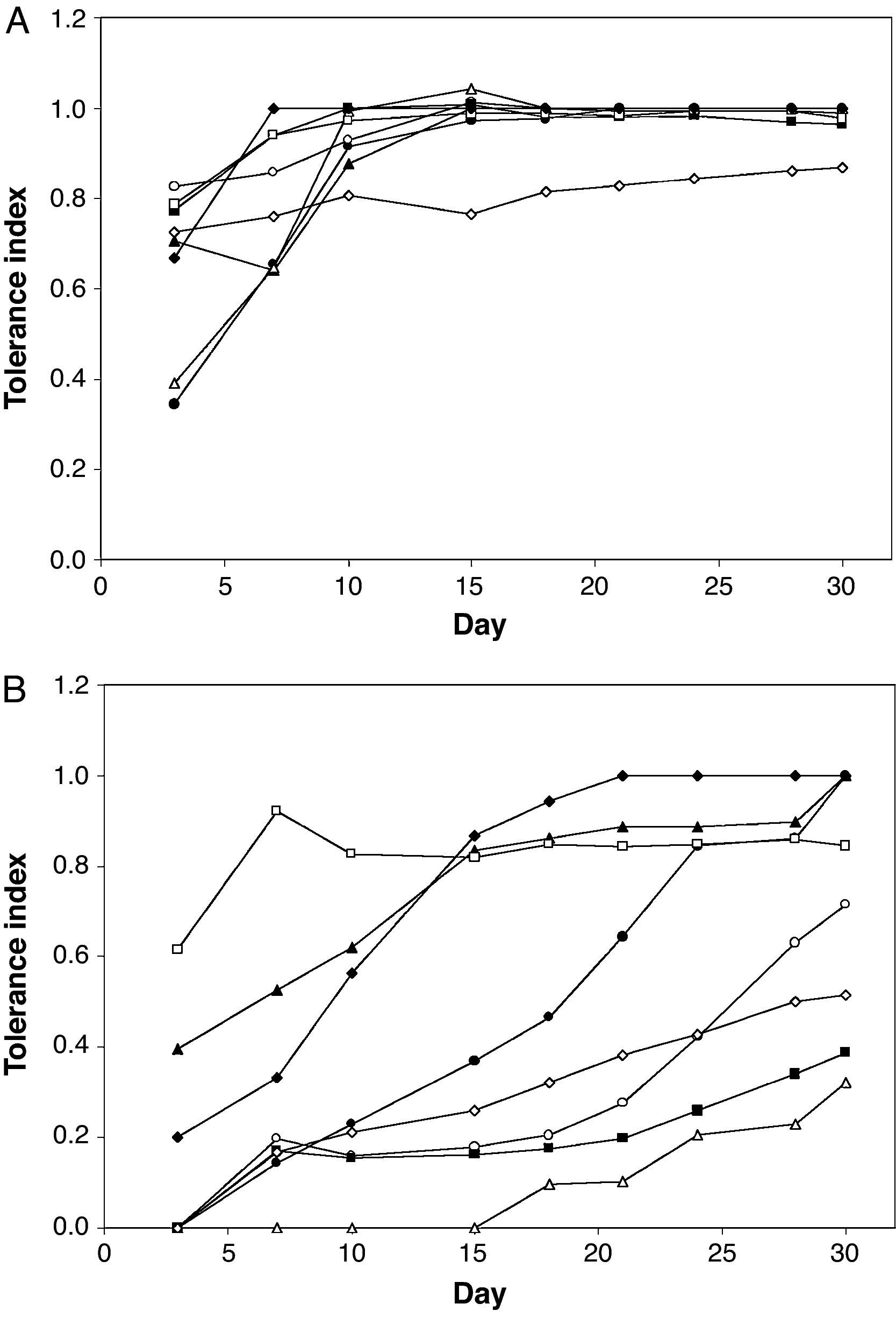
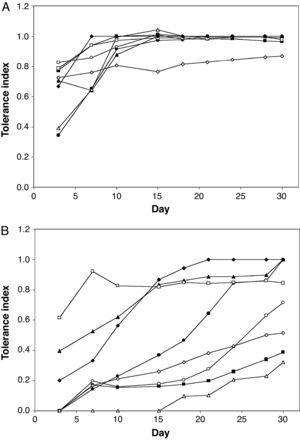
Figure 2 shows the tolerance index to ACzA and 90% AASW for eight mould isolates from the AAS. From day 15 on, all isolates except Eupenicillium sp. RA403 showed a tolerance index to ACzA that was close to one, regardless of their initial tolerance index (measured on day 3). On the contrary, the tolerance index for 90% AASW medium was much more variable. Three groups of isolates could be distinguished by growth at day 30: (1) isolates with a tolerance index equal to one (RA101, RA207 and RA402); (2) isolates with a tolerance index less than 0.5 (RA208 and RA302); and (3) isolates with a tolerance index in the range 0.5-1.0 (RA102, RA306 and RA403). Although most isolates showed a general trend to increase their tolerance index through the experiment, the magnitude of such an increase was variable. Further variability was observed in the delay of growth, defined as the time elapsed from inoculation on selective medium to the moment when the fungus started to grow and extend to the rest of the plate. The extreme case was represented by Trichoderma sp. RA208, which showed delayed growth on 90% AASW medium for more than 15 days.
Tolerance of eight mould isolates from the AAS to ACzA (A) and 90% AASW medium (B).
(¿) Paecilomyces sp. RA101; (○) Penicillium sp. RA102; (▴) Aspergillus section Fumigati RA207; (▵) Trichoderma sp. RA208; (¿) Penicillium sp. RA302; (□) Penicillium sp. RA306; (♦) Aspergillus section Nigri RA402; (¿) Eupenicillium sp. RA403.
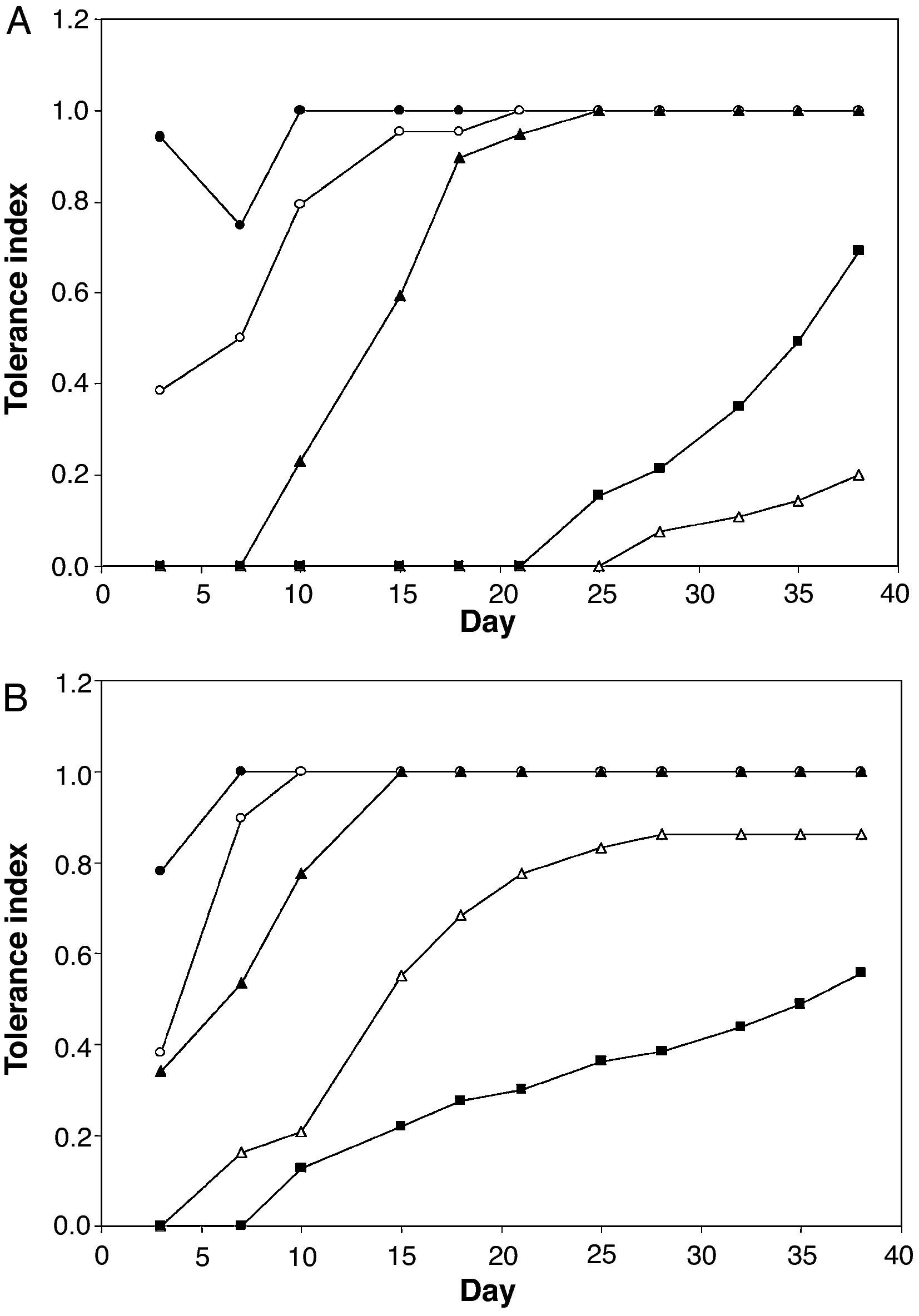
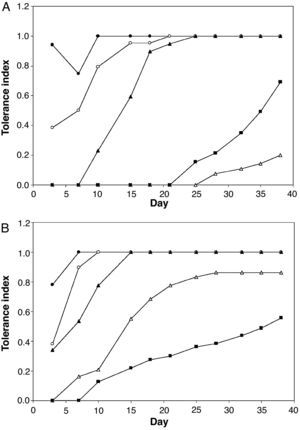
Table 1 shows the minimum inhibitory concentration of AASW for the two Aspergillus collection strains. None germinated in media with a high concentration of AASW after inoculation of 106 conidia. However, both strains grew on media containing up to 90% AASW when a small mycelial disk cut from sporulated cultures on non-selective medium was used as an inoculum. Figure 3 shows the tolerance indexes for these two strains at different concentrations of AASW over 38 days. The data suggest that the tested A. niger strain was more tolerant to AASW than the A. fumigatus strain as, in general, it showed greater tolerance index values at different AASW concentrations, and a shorter delay in growth when cultured on media with more than 30% AASW. The A. fumigatus collection strain had an unexpected response because, during all the experiment, its tolerance index for 90% AASW medium was higher than its tolerance index for 70% AASW medium. Moreover, its tolerance index for 90% AASW medium at the end of the experiment was even higher than that of the collection strain of A. niger.
After studying the trend of the tolerance index to 90% AASW medium, only Penicillium sp. RA306 and Aspergillus section Nigri RA402 out of the 10 mould isolates used (eight from the AAS and two from our culture collection) germinated in that selective medium. These results are in agreement with those obtained when spores from Sabouraud's agar cultures of these 10 isolates were used to inoculate 90% AASW medium.
After three days of incubation, all R. toruloides isolates and the C. ibericus isolate from the AAS formed colonies on ACzA and 90% AASW medium. In contrast, the five environmental isolates of A. pullulans and the two yeast strains from our culture collection showed a similar behaviour to most mould isolates, forming visible colonies on the control medium and ACzA but not on 90% AASW medium. The minimum concentration of AASW that inhibited the growth is presented for some isolates in Table 1.
DiscussionThe IPB contains interesting examples of extreme environments, where two factors traditionally considered as limiting for life coexist: extremely low pH and high levels of metals. In spite of these harsh conditions, life in these environments is not only possible, but an astonishing diversity of bacterial, fungal, algal and protistan species thrive in them.1–5,10–12,22–23
The microbial communities that inhabit the AAS have not been as extensively studied as those in nearby acidic environments, such as the Tinto and Odiel rivers. In fact, to date, only a first characterization of the phytoplankton community at a point near the Tharsis mine has been performed.26 In this work we studied the fungal community at the same place.
Our results suggest that low pH is not a limiting factor for the development of fungi in the AAS, as all isolates studied were able to grow when cultured on medium adjusted with HCl. These results are in agreement with the field observations of Aguilera et al.,1 who, after studying different physicochemical parameters and the composition of benthic communities of eukaryotic microbes at 12 sampling points along the Tinto River, concluded that the distribution of those communities was more influenced by the presence of heavy metals than by pH.
Nonetheless, our results show that not all fungi that inhabit an extreme environment are equally adapted to it. Only four out of the 18 mould isolates from the AAS were able to germinate on 90% AASW medium. Moreover, differences in the growth pattern, final tolerance index value, and delay of growth were observed among some isolates when mycelial disks from cultures on Sabouraud agar were used as an inoculum.
The yeast species isolated from the AAS were R. toruloides and C. ibericus, which were able to grow on 90% AASW medium. Yeasts of the genera Rhodosporidium and Cryptococcus have been found in other extreme acidic environments, not only within the IPB,10,11,23 but also in other parts of the world.30,31
Five isolates of the dimorphic fungus A. pullulans were also recovered from the AAS, none of which formed visible colonies on 90% AASW medium. Moreover, the minimum inhibitory concentrations of AASW for two isolates of this fungus were similar or lower than those obtained for collection strains.
The inability of some fungal isolates from extreme environments to grow in media that simulate their environmental conditions has been previously reported. For example, López-Archilla et al.23 observed that 48% of the yeast and 56% of the moulds isolated from the Tinto River were unable to grow in a liquid medium prepared with water from that river. The presence of fungi that are unable to tolerate the extreme environmental conditions of these habitats might be the result of dispersal from mesophilic areas. However, the possibility that fungi are unable to grow in culture media prepared with water from these extreme environments, but can grow in their natural habitat cannot be excluded.
In both the Tinto River and in the AAS, the microbial community mainly forms biofilms that are distributed along the river bed. In these formations, the entire community is usually embedded in a mucilaginous coating that probably changes the physicochemical conditions within the biofilm, protecting microorganisms from the extremely acidic conditions and the high concentrations of metals present in water.1,3 In fact, different mechanisms of tolerance or resistance to multiple metals have been described for biofilm microorganisms.16 Furthermore, the possibility that other factors, such as seasonal variations in the physicochemical conditions of the environment, or interaction with other microbial species, may affect the ability of the different fungal isolates to grow in natural conditions should not be ignored.
In general, most of the fungal taxa isolated in this work from the AAS are widely distributed in the environment. The presence of eukaryotic microorganisms closely related to neutrophiles in environments affected by acid mine drainage (AMD) has been highlighted by other authors.6,7 This fact could be interpreted as a recent colonization of low-pH, metal-rich extreme environments by neutrophilic lineages of eukaryotes.6,7 In constrast, most prokaryotic members of AMD communities form distinct lineages of strict acidophiles, suggesting an early colonization of their extremely harsh habitat.6,7 Alternatively, some eukaryotic microorganisms may have traits that provide them with a selective advantage for survival in acidic, metalliferous environments.6,7 To test this possibility, we studied the ability of different culture collection strains of moulds and yeasts to grow on AASW medium.
The results of this work demonstrate that fungal collection strains can grow on media prepared with high concentrations of AASW. These strains, however, showed an increased delay of growth when cultured on selective media and reached a lower tolerance index than Aspergillus isolates from the AAS. The greater delay in growth of these strains in AASW media may be due to the lack of a pre-adaptation step to the stressors found at the AAS. Moreover, differences in ability to germinate were observed between the A. niger collection strain and the environmental Aspergillus section Nigri isolates; while conidia of the collection strain germinated only at relatively low concentrations of AASW of up to 40% (v/v), the environmental isolates germinated on 90% AASW medium.
In contrast to mould collection strains, none of the tested yeast collection strains formed visible colonies on 90% AASW medium. However, the possibility that other collection strains might grow on this selective medium cannot be eliminated.
Since fungi play an important role as decomposers in ecosystems, their tolerance to environmental stressors is relevant from an ecological point of view. The adaptation of fungi and other microorganisms to metal-rich environments might be the result of different physiological mechanisms, such as bioabsortion, bioprecipitation, extracellular sequestration, and mechanisms of transport and/or chelation.15 Furthermore, several investigations have demonstrated that the adaptation of some eukaryotic microorganisms to extreme environments, including the AAS, may be due to preadaptive mutations and the subsequent selection of resistant mutants.8,9,26 However, the role of preadaptive mutation in fungal adaptation to extreme environments has not yet been explored.
In conclusion, the AAS is an extreme environment that harbours fungal species that manifest differences in their growth pattern when cultured on media that simulate the acidic, metal-rich conditions of their natural habitat. Continued investigation is needed to understand the genetic and/or physiological mechanisms that allow fungi to inhabit this extreme environment.
FinancingThis work was supported by a research grant from the Complutense University and the Madrid Autonomous Community (GR85/06). Sergio Álvarez-Pérez acknowledges a grant from the FPU programme (ref. AP 2005-1034), Spanish Ministry of Education and Science.
Conflict of interestThe authors report no conflict of interest.
Eduardo Costas and Victoria López-Rodas are gratefully acknowledged for providing the environmental samples. The authors also appreciate the constructive comments of an anonymous reviewer.