The best laboratory diagnostic approach to detect Clostridioides [Clostridium] difficile infection (CDI) is a subject of ongoing debate. With the aim of evaluating four laboratory diagnostic methods, 250 unformed stools from patients with suspected CDI submitted to nine medical center laboratories from November 2010 to December 2011, were studied using: (1) an immunochromatographic rapid assay test that combines the qualitative determination of glutamate dehydrogenase (GDH) plus toxins A and B (QAB), the CDIFF QUIK CHEK COMPLETE assay; (2) an enzyme immunoassay for qualitative determination of toxins A and B, the RIDASCREEN™ C. difficile Toxin A/B assay (RAB); (3) a PCR for the toxin B gene assay (PCR); and (4) the toxigenic culture (TC). C. difficile isolates from direct toxin negative stools by QAB, RAB and PCR were evaluated for toxigenicity by the same direct tests, in order to assess the contribution of the TC (QAB-TC, RAB-TC, PCR-TC). A combination of the cell culture cytotoxicity neutralization assay (CCCNA) in stools, and the same assay on isolates from direct negative samples (CCCNA-TC) was considered the reference method (CCCNA/CCCNA-TC). Of the 250 stools tested, 107 (42.8%) were positive by CCCNA/CCCNA-TC. The GDH and PCR/PCR-TC assays were the most sensitive, 91.59% and 87.62%, respectively. The QAB, RAB, QAB/QAB-TC and RAB/RAB-TC had the highest specificities, ca. 95%. A negative GDH result would rule out CDI, however, its low positive likelihood ratio (PLR) of 3.97 indicates that a positive result should always be complemented with the detection of toxins. If the RAB, QAB, and PCR assays do not detect toxins from direct feces, the toxigenic culture should be performed. In view of our results, the most accurate and reliable methods to be applied in a clinical microbiology laboratory were the QAB/QAB-TC, and RAB/RAB-TC, with PLRs >10 and negative likelihood ratios <0.30.
El mejor procedimiento para realizar el diagnóstico de laboratorio de la infección causada por Clostridioides [Clostridium] difficile (ICD) es aún objeto de debate. Con el fin de evaluar cuatro métodos diagnósticos de laboratorio, se estudiaron 250 muestras de heces diarreicas provenientes de pacientes con sospecha de ICD remitidas a los laboratorios de nueve centros médicos entre noviembre de 2010 y diciembre de 2011. Dichas muestras se analizaron mediante los siguientes métodos: 1) un ensayo rápido inmunocromatográfico que combina la detección cualitativa de la glutamato deshidrogenasa (GDH) y de las toxinas A y B (QAB), CDIFF QUIK CHEK COMPLETE; 2) un enzimoinmunoanálisis para la determinación cualitativa de las toxinas A/B, RIDASCREEN™ C. difficile Toxin A/B (RAB); 3) un método molecular basado en PCR para la detección del gen que codifica la toxina B (PCR) y 4) el cultivo toxigénico (TC). Como método de referencia se utilizó la combinación del ensayo de citotoxicidad sobre cultivo de células con la neutralización de toxina mediante anticuerpo específico en los filtrados de las heces (CCCNA) y el mismo método en sobrenadantes de aislamientos de C. difficile (CCCNA-TC). La toxigenicidad de las cepas aisladas de muestras directas negativas con QAB, RAB y PCR se evaluó con los mismos métodos, con el propósito de detectar la contribución del TC (QAB-TC, RAB-TC, PCR-TC). De las 250 muestras estudiadas, 107 (42,8%) fueron positivas por CCCNA/CCCNA-TC. Los métodos GDH y PCR/PCR-TC fueron los más sensibles: 91,59 y 87,62%, respectivamente. Los métodos QAB, RAB, QAB/QAB-TC y RAB/RAB-TC mostraron las mayores especificidades, del 95%, aproximadamente. Un resultado negativo para GDH excluiría la ICD, pero su baja razón de verosimilitud positiva (PLR), que fue 3,97, indica que un resultado positivo debe complementarse con la detección de toxinas. Cuando no se detectan toxinas directas por RAB, QAB ni PCR, debería realizarse el TC. De acuerdo con nuestros resultados, los métodos más precisos y confiables para ser aplicados en un laboratorio de microbiología clínica son QAB/QAB-TC y RAB/RAB-TC, con una PLR>10 y una razón de verosimilitud negativa<0,30.
Clostridioides [Clostridium] difficile is increasingly recognized as an important nosocomial pathogen with significant morbidity and mortality. C. difficile infection (CDI) is a serious healthcare concern, with high incidence and recurrence rates, healthcare costs, and which is sometimes refractory to standard treatment3,9,22,23,38,43,56,57.
C. difficile is known to express up to three toxins: toxin A (TcdA), toxin B (TcdB), and 6–12.5% of strains produce a third toxin called binary toxin (CDT). These toxins cause extensive colonic inflammation, epithelial tissue damage, and cell death. CDI shows a wide spectrum of clinical manifestations, from a mild, self-limited diarrheal illness to a fulminant, life-threatening colitis and recurrence. Its acquisition is mostly related to hospital or healthcare, however, it can also be observed in outpatients. Moreover, it is important to note that community-associated CDI (CA-CDI) may occur in individuals who lack the traditional risk factors such as antibiotic use, advanced age, and severe underlying disease8,30,68. In the past decade, the epidemiology of C. difficile changed and new types have emerged7,23.
C. difficile is genetically heterogeneous, as well as its geographical distribution. Outbreaks related to strains that produce the binary toxin, called “hypervirulent” types (ribotype 027 and ribotype 078), have been detected in some countries and associated with more severe disease. The 027/NAP1/BI strain also carries an 18-bp deletion and a 1-bp deletion (at nucleotide [nt] 117) in the tcdC gene, a putative negative regulator of tcdA and tcdB gene expression1,8,28. Several pathogenic TcdA-negative, TcdB-positive strains that have been found in Asia and Latin America – including Argentina – may affect the CDI diagnosis depending on the kind of tests performed5,14,16,29,34,37,39–41,44.
The best laboratory diagnostic approach to detect CDI is a subject of ongoing debate4,25,67.
Since C. difficile colonization or infection cannot be distinguished by laboratory testing, toxin detection should be performed only in symptomatic patients. It consists of detecting toxigenic C. difficile or its toxins A and B in unformed stools, when illness due to C. difficile is suspected4. Historically, the cell culture cytotoxicity neutralization assay (CCCNA) was considered the gold standard method for the diagnosis of CDI. However, many laboratories do not have the technical expertise, facilities or training to perform it. In addition, it requires at least 16h to obtain the result. Toxins A/B enzyme immunoassay (EIA) is the most widely used test for the detection of C. difficile TcdA and TcdB, but it is associated with poor sensitivity, thus with an unacceptable high rate of false-negative results20,47,55.
Polymerase chain reaction (PCR), nested PCR and real-time PCR have been found to be effective for the detection of toxin genes in stool specimens, with good sensitivity and specificity13,15,24,33,36,50,63.
The detection of toxigenic strains of C. difficile is currently a more sensitive approach for the diagnosis of CDI; however, the culture is infrequently done for diagnostic purposes because of its slow turnaround time, the need for special isolation media and technical expertise. On the other hand, the toxigenic culture (TC) is recommended as the gold standard method because it is the best performing test available. Indeed, it shows better sensitivity (90–100%), and specificity (98–100%) than the traditionally used CCCNA (70–100% and 90–100%, respectively). Moreover, TC is essential for further epidemiological and susceptibility studies12,46,54.
On the other hand, the glutamate dehydrogenase (GDH) assay, which detects the antigen present in both toxigenic and nontoxigenic strains of C. difficile, and directly in stool samples, is simple, rapid and appears to have good sensitivity. However, it lacks specificity, and like the culture, needs to be followed by toxin detection26,58–60.
The use of the GDH assay as screening and the confirmation of its positive results with the detection of TcdA and TcdB by EIA, TC or CCCNA seems to be the best algorithm to perform in laboratories. The nucleic acid amplification tests, especially PCR assay for the detection of C. difficile toxin B gene (tcdB), have emerged as an option of a multitest algorithm12,35.
The purpose of this study was to assess the usefulness of four available laboratory methods for the diagnosis of CDI. An immunochromatographic rapid assay test that combines the qualitative determination of toxins A and B of C. difficile plus GDH in stool samples, an enzyme immunoassay for the qualitative determination of C. difficile toxins A and B in human stool specimens, a PCR assay to detect the tcdB toxin gene of C. difficile, and the toxigenic culture were performed. The combination of a direct cytotoxicity assay from stool samples and the cytotoxicity assay from stool isolates results was considered the gold standard method.
Materials and methodsFrom November 2010 to December 2011, 250 unformed fecal samples of adult patients at risk of CDI were provided for the study by the following laboratories (n;%): Sanatorio de la Trinidad Mitre, Ciudad Autónoma de Buenos Aires (CABA) (49; 19.6); Hospital General de Agudos Dr. Enrique Tornú, CABA (34; 13.6); Instituto de Investigaciones Médicas Alfredo Lanari – Universidad de Buenos Aires, CABA (30; 12); Hospital de Clínicas José de San Martín – Universidad de Buenos Aires, CABA (30; 12); Hospital de Infecciosas Dr. Francisco J. Muñiz, CABA (25; 10); Hospital Nacional de Clínicas de Córdoba, Córdoba (25; 10); Hospital Interzonal General de Agudos Presidente Perón, Avellaneda, Provincia de Buenos Aires (20; 8); Hospital de Emergencias Clemente Álvarez, Rosario (HECA), Provincia de Santa Fe (20; 8); and Hospital Alemán, CABA (17; 6.4). Stool specimens were submitted in sterile containers and stored at 4°C up to 24h. Prior to process, samples were mixed well and split using a sterile disposable graduated pipette. A 0.5ml aliquot of stool specimen was transferred from the original collection container to a sterile cryovial and frozen at −20°C for further PCR. A 1ml aliquot of stool was added to 1ml of saline solution (NaCl 0.085g/l), homogenized vigorously and centrifuged at 3000×g for 15min. The supernatant was filtered through a 0.45μm membrane and the fecal filtrate was transferred to a cryovial and frozen at −20°C for further CCCNA. A 0.5ml aliquot was used for the enzyme immunoassays, and a 0.5–1ml aliquot of stool sample was transferred to a sterile glass test tube for culture. Stool specimens were tested by QAB, RAB, PCR and CCCNA. An ethanolic shock was performed prior to the stool culture. The toxigenic isolates were characterized using standard phenotypic methods61.
C. difficile colonies were suspended in brain heart infusion (BHI) broth (Britania™, Argentina) and incubated in anaerobic conditions. An aliquot was centrifuged for 5min at 13000×g and cell free supernatants (CFS) were obtained by filtration as described below for further CCCNA-TC.
In order to assess the contribution of the TC, C. difficile isolates from toxin-negative stool specimens by QAB, RAB, PCR and CCCNA were tested for toxigenicity by the same direct tests named QAB-TC, RAB-TC, PCR-TC, and CCCNA-TC. The combination of toxin detection results in supernatants from stool specimens (CCCNA) and from isolates (CCCNA-TC) was used as the reference test (CCCNA/CCCNA-TC).
Combination testing – GDH detection and toxin EIA. C DIFF QUIK CHEK COMPLETE™ (Techlab, Inc., USA)It is a rapid cassette membrane enzyme immunoassay for the simultaneous detection of the GDH antigen and TcdA and TcdB of C. difficile in fecal specimens. Testing was performed on stool samples and their isolates according to the manufacturer's recommendations. The C Diff Quik Chek Complete test detects the GDH antigen of C. difficile and toxins A and B, without differentiation of the toxins. With the purpose of providing most useful information of the results of the GDH assay alone, we evaluated its detection independently (Table 1).
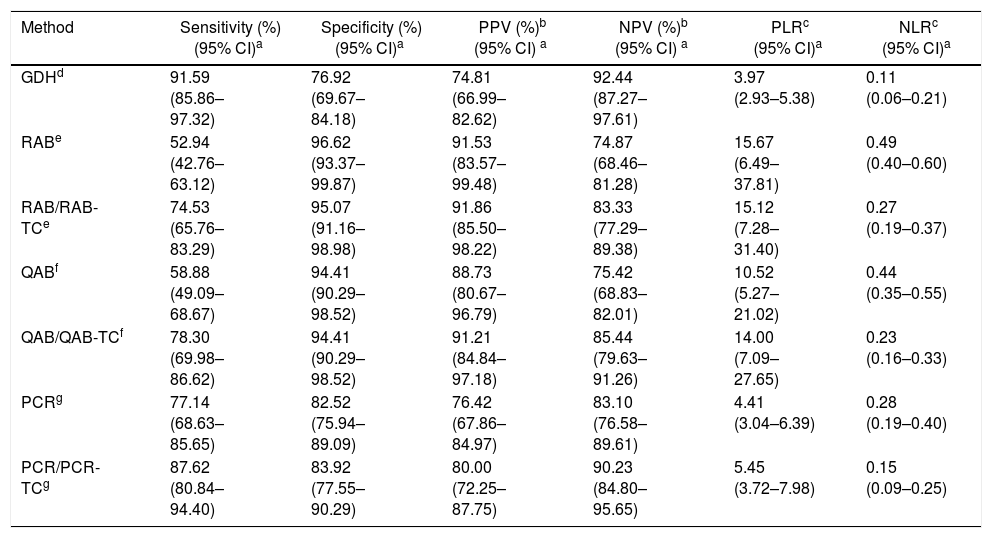
Comparison of Clostridioides [Clostridium] difficile diagnostic testing directly from stool samples and from the stool isolates, against the results of the combination of CCCNA/CCCNA-TC
| Method | Sensitivity (%) (95% CI)a | Specificity (%) (95% CI)a | PPV (%)b (95% CI) a | NPV (%)b (95% CI) a | PLRc (95% CI)a | NLRc (95% CI)a |
|---|---|---|---|---|---|---|
| GDHd | 91.59 (85.86–97.32) | 76.92 (69.67–84.18) | 74.81 (66.99–82.62) | 92.44 (87.27–97.61) | 3.97 (2.93–5.38) | 0.11 (0.06–0.21) |
| RABe | 52.94 (42.76–63.12) | 96.62 (93.37–99.87) | 91.53 (83.57–99.48) | 74.87 (68.46–81.28) | 15.67 (6.49–37.81) | 0.49 (0.40–0.60) |
| RAB/RAB-TCe | 74.53 (65.76–83.29) | 95.07 (91.16–98.98) | 91.86 (85.50–98.22) | 83.33 (77.29–89.38) | 15.12 (7.28–31.40) | 0.27 (0.19–0.37) |
| QABf | 58.88 (49.09–68.67) | 94.41 (90.29–98.52) | 88.73 (80.67–96.79) | 75.42 (68.83–82.01) | 10.52 (5.27–21.02) | 0.44 (0.35–0.55) |
| QAB/QAB-TCf | 78.30 (69.98–86.62) | 94.41 (90.29–98.52) | 91.21 (84.84–97.18) | 85.44 (79.63–91.26) | 14.00 (7.09–27.65) | 0.23 (0.16–0.33) |
| PCRg | 77.14 (68.63–85.65) | 82.52 (75.94–89.09) | 76.42 (67.86–84.97) | 83.10 (76.58–89.61) | 4.41 (3.04–6.39) | 0.28 (0.19–0.40) |
| PCR/PCR-TCg | 87.62 (80.84–94.40) | 83.92 (77.55–90.29) | 80.00 (72.25–87.75) | 90.23 (84.80–95.65) | 5.45 (3.72–7.98) | 0.15 (0.09–0.25) |
It is an EIA for the qualitative determination of the C. difficile toxins A/B in stool specimens and from cultures of toxin-producing C. difficile strains. The test was performed according to the manufacturer's instructions on stool specimens and stool isolates.
Cell-culture cytotoxicity neutralization assayFecal filtrates and CFS were stored at −20°C until CCCNA/CCCNA-TC were performed. Cell cultures: Vero cells were grown in Dulbecco's modified Eagle's medium (DMEM; Gibco BRL Life Technologies, Rockville, MD, USA) supplemented with 10% (v/v) inactivated fetal calf serum (30min, 60°C) (BIOSER, Argentina, PAA Laboratories GmbH), 2g NaCO3H, streptomycin 10mg/l and penicillin G 10IU/ml. Cells (4×104 cells per well) were inoculated in 96-well tissue culture plates (Corning, NY, USA) and incubated at 37°C for 48h in a 5% CO2/95% air atmosphere. Cytotoxicity assay: cultured cells were washed twice with 0.2ml of phosphate-buffered saline (PBS; KH2PO4 0.144g/l, NaCl 9g/l, Na2HPO4 0.795g/l, pH 7.5). One hundred microliters of two-fold dilutions of fecal filtrates or CFS in DMEM (without fetal calf serum) were added per well and incubated at 37°C for 16h in a 5% CO2/95% air atmosphere. Biological activity through determination of cytopathic effects (cell rounding) was evaluated by microscopy. Positive samples were confirmed when cytopathic effect was neutralized in the presence of anti-TcdB monoclonal antibody (Meridian Life Science, Inc., USA). The titer of biological activity was defined as 1/(sample dilution) that produces rounding on 100% of cells45,62,67.
Polymerase chain reactionThe PCR was performed in previously frozen fecal samples and suspensions of isolates in sterile distilled water stored at −20°C. DNA was extracted from 200mg of the stool by using QIAamp DNA Stool Kit (Qiagen, Germany) according to the manufacturer's recommendations. The final volume of the DNA extract was 200μl. DNA was extracted from 2 to 3 C. difficile colonies that were suspended in ultrapure distilled water, then boiled for 10min and centrifuged at 15000×g for 2min. PCRs were carried out in 25μl of volume reaction, where 6.25μl of the extract was added to 18.75μl of the reaction mix consisting of: 2.5mM MgCl2, 50mM KCl, 10mM Tris–HCl (pH 9.0), the four deoxynucleoside triphosphates (0.075mM of each other), 0.4μM of each primer, and 0.05U/μl of Taq DNA polymerase. A segment of the tcdB gene was amplified by using primer NK104-sequence 5′-GTGTAGCAATGAAAGTCCAAGTTTACGC-3′; positions 2945 to 2972- and primer NK105 -sequence 5′-CACTTAGCTCTTTGATTGCTGCACCT-3′; positions 3123 to 3148-, which were derived from the nonrepeating sequence of the tcdB gene of C. difficile32,64. The thermal profile was one cycle at 95°C for 4min followed by 35 cycles comprising one step at 95°C for 20s, 60°C for 20s and 72°C for 20s. At the end of the PCR cycles, the tubes were incubated at 72°C for 7min. The presence of a 204-bp fragment was considered indicative of the presence of the tcdB gene. The amplified product was analyzed until separation by electrophoresis on 2% agarose gel with ethidium bromide. A positive control of pure DNA from toxigenic C. difficile strain ATCC 43255, a negative control of pure DNA from a clinical isolate of Clostridium sordellii, and sterile distilled water were added to each run. For the detection of inhibitors, 2μl of each eluate extracted from all PCR-negative samples was spiked with 2μl of the eluate from a PCR-positive stool.
Ethanolic shock of stools and cytotoxigenic culturesAll stool specimens were subjected to ethanolic shock with 1:1vol/vol absolute ethanol/stool sample, mixed and incubated for 30min at room temperature. Then, they were cultured on brain heart infusion agar (BHIA) supplemented with 5% lysed horse blood, and on BHIA with 16mg/l of cefoxitin (BHIAC). Plates were incubated anaerobically at 37°C and examined after 48–72h. Gray-white colonies with irregular filamentous edges or opaque appearance on BHIA or BHIAC plates, morphologically resembling C. difficile, were Gram-stained. If suspicious for Clostridium spp., they were subcultured onto Brucella blood agar plates (Britania™) and also tested for aerotolerance on chocolate agar plates (Britania™) in a 5% CO2 atmosphere. C. difficile was identified by no growth at 37°C after 48–72h of incubation under 5% CO2, a positive result for the proline aminopeptidase test, gelatin and esculin hydrolysis, a negative result for indole, lecithinase and lipase production, and having a characteristic “horse barn” odor. C. difficile isolates that grew on BHIAC or BHIA from toxin-negative stool samples were tested for toxigenicity. Isolates were inoculated into BHI broth and incubated in anaerobic conditions. They were centrifuged at 3000×g for 10min, and the supernatants were tested by the same direct methods CCCNA, EIAs, and PCR.
Statistical analysisSensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), positive likelihood ratio (PLR) and negative likelihood ratio (NLR) with their corresponding 95% confidence intervals were calculated for all the assays. Because PPVs and NPVs depend on the prevalence of C. difficile, we evaluated the clinical diagnostic value of the different tests by comparing the results obtained with the likelihood ratios (NLR and PLR) to become independent of those parameters. All parameters were estimated and analyzed by the SPSS Statistics version 20.0 software (SPSS, Inc., Chicago, IL).
ResultsOf the 250 fecal samples studied, 107 (42.8%) were positive by CCCNA/CCCNA-TC.
The performance of the GDH, QAB, RAB, and PCR assays in stools, and the QAB-TC, RAB-TC and PCR-TC assays on isolates was assessed as follows: all 250 samples were tested by the RAB, QAB, and GDH assays; 249 samples were available to be tested using the QAB-TC, 248 using RAB-TC and PCR-TC, and 247 by PCR assays.
The results of each method were compared against the results of the CCCNA/CCCNA-TC combination. Sensitivity, specificity, PPV, NPV, PLR, and NLR of the GDH, QAB, RAB, PCR, QAB/QAB-TC, RAB/RAB-TC and PCR/PCR-TC assays and their 95% confidence interval are summarized in Table 1, which shows the results of the comparison of C. difficile detection methods with the results of the gold standard CCCNA/CCCNA-TC.
The highest sensitivities and NPVs were found with GDH and PCR/PCR-TC, approximately 90% with both methods. Specificities and PPVs were high, approximately 95% and approximately 90% with RAB, RAB/RAB-TC, QAB, and QAB/QAB-TC, respectively.
Of the 107 positive samples by the gold standard, 47 (43.93%) and 7 (6.54%) specimens were determined to be positive and negative by all assays, respectively. The GDH and PCR were the only positive tests in 6/107 (5.6%), and 1/107 (0.9%) positive samples, respectively.
Accordingly, RAB, RAB/RAB-TC, QAB and QAB/QAB-TC assays with PLRs >10, and GDH, PCR/PCR-TC, PCR, QAB/QAB-TC and RAB/RAB-TC assays with NLRs <0.30 demonstrated to be useful to be applied in a clinical microbiology laboratory; however, the most accurate and reliable methods for diagnosis of CDI proved to be the QAB/QAB-TC or RAB/RAB-TC with both, PLRs >10 and NLRs <0.30.
DiscussionDespite the availability of several laboratory methods to diagnose CDI, the optimal strategy is debatable6. The guidelines recommend the implementation of a multitest algorithm that uses GDH assays to screen for C. difficile in stool specimens, followed by either, CCCNA and/or TC to identify toxin-producing C. difficile strains. However, these procedures are not so useful in the clinical setting due to slow time to results, labor-intensity and need for special laboratory facilities. GDH assays have high sensitivity and NPV, but cannot differentiate between toxigenic and nontoxigenic strains. Therefore, they must be used sequentially with toxin confirmatory testing. Tox A/B EIAs that could be used alone or in combination with GDH are fast and specific for toxin detection, but limited because of their low sensitivity. PCR-based toxin gene testing is also fast; however, their usefulness in the diagnosis of CDI is controversial12,15.
Thus, the best testing strategy to diagnose CDI in the clinical laboratory to provide timely, cost-effective, and accurate results remains a subject of controversy. It is important to take into account that, in this study, the sensitivity of the assays was determined by comparing the results to those of CCCNA/CCCNA-TC. Since TC has better sensitivity than the CCCNA for detecting toxigenic C. difficile strains, it is not surprising that the sensitivities reported by others when comparing the assays to the gold standard (TC) are lower than those reported with CCCNA as the reference method15,19.
Glutamate dehydrogenaseGDH, encoded by the gluD gene, is a metabolic enzyme produced by both toxigenic and nontoxigenic C. difficile strains and other Clostridium spp.; thus, its diagnostic utility is as a screening marker10. In the present study, the detection of GDH had the highest sensitivity (91.59%). The NLR of the GDH was 0.11, that is low enough to indicate that a negative result would rule out the CDI. However, it had the lowest specificity (76.92%) and PLR (3.97), indicating that a positive result should always be complemented with the detection of toxins or their encoding genes. Moreover, many reports point out differences in GDH assay sensitivities; indeed, this approach remains an interim recommendation in the Society for Healthcare Epidemiology of America (SHEA) Guidelines12. Although our study and early studies comparing the GDH assay to CCCNA demonstrated high sensitivity and negative predictive values and concluded that the test for GDH is excellent for screening, more recent comparisons to TC and PCR have shown a sensitivity of 71–88%, which puts into question the value of these algorithms11,48,49. These different sensitivities might be possibly due to regional/geographical differences in strain ribotypes but also to different detection methods that could affect the GDH assays12,27. In this study, we demonstrated an 8.41% rate of false negatives, showing a good test performance.
Enzyme immunoassaysIn the present study, RAB and QAB showed the lowest sensitivity – 52.94% and 58.88%, respectively; and the highest specificity – 96.62 and 94.41, respectively. These results agree with those reported by others2,21,25. As noted, the RAB performance was comparable to that of the QAB with a slight decrease in the sensitivity test and a slight increase in the specificity test. Our data demonstrated that the addition of the EIA toxin detection on C. difficile isolates from direct toxin-negative stools (QAB-TC and RAB-TC), improved the sensitivity of the EIA kits of each single test (QAB and RAB). Indeed, the RAB/RAB-TC and the QAB/QAB-TC were the best toxigenic C. difficile detection methods evaluated, with sensitivity, specificity, PLR and NLR of 74.53%, 95.07%, 15.12, 0.27 and 78.30%, 94.41%, 14 and 0.23, respectively. Their good PLRs indicate that patients with CDI have a very high chance to show positive tests compared with non-CDI subjects.
Polymerase chain reactionAlthough the PCR assay seems to be rapid, sensitive and specific, its routine use is not recommended until more data are available12.
In this study, we compared a PCR and a PCR/PCR-TC assay to detect the tcdB gene against CCCNA/CCCNA-TC. Ours results demonstrated that PCR assays may serve as suitable methods only for excluding but not for confirming CDI, conversely to those reported by Deshpande et al. in their meta-analysis on PCR assay performances, with overall means of sensitivity, specificity, PLR, and NLR of about 90%, 96%, 26.89 and 0.11, respectively15. The low specificity and PLR shown in this study are clinically undesirable for a diagnostic assay, because a false positive result may lead to unnecessary antimicrobial treatment and isolation of the patient19.
Several factors could have contributed to the differences observed in the results from the literature. It is possible that inter-study differences in performing the techniques used have occurred. It is important to take into account that most published data were obtained with real-time PCR assays instead of with a conventional PCR assay. Unlike conventional PCR, real-time PCR is more sensitive and has better resolution. Moreover, since there is no accepted real gold standard, CCCNA or TC are often used as reference tests15. Thus, it is possible that the use of a different reference standard – a combination of CCCNA and CCCNA-TC – could lead to an underestimation of the sensitivities and specificities obtained. This study reinforces the concept that non-commercial molecular methods can only be used for diagnostic purposes once validated by the reference method.
There are some potential limitations of using PCR as a diagnostic tool. It detects the tcdB gene encoding the toxin and not the toxin itself. Therefore, it might detect colonization instead of infection31,66. In fact, other authors have reported that reliance on molecular testing instead on toxin testing can result in overdiagnosis and overtreatment of CDI17,51. In view of these observations, it seems prudent not to recommend the PCR assay until more data are available regarding the diagnosis of CDI, outcome, and response to specific antimicrobial therapy.
It seems interesting to comment that diarrhea has many causes in hospitalized patients, where many of whom could be colonized with C difficile strains, especially if they were staying in long-term care facilities, where C. difficile is the cause of diarrhea in 5% up to 30% of hospitalized patients who have diarrhea and are tested for C. difficile12,42.
While a diagnostic assay may indicate the presence or absence of the organism or its toxins, the test by itself does not determine who does or does not have CDI. Thus, it is important to use test methods that can help in this matter because an accurate clinical diagnostic is mandatory17,18,52,53. Moreover, it should be remembered that the diagnosis of CDI could be missed in patients whose stools are screened only for the presence of toxins5,29,34,40,41,65.
In summary, despite the presence of toxigenic C. difficile, approximately 6% of samples may be negative by all the methods. When the rest of the tests are negative, the GDH, and PCR may be the only positive assays (5.6% and 0.9%, respectively). The GDH assay appeared to be a good screening test to rule out negative specimens and to select the samples for further testing. The GDH-positive stools must be confirmed using toxin tests to distinguish between toxigenic and nontoxigenic strains. Depending on the available possibilities of the laboratory and in view of the results, an appropriate strategy could be the use of TOX A/B EIAs in stools, followed by culture plus toxin detection from the isolates by EIAs, PCR or CCCNA, in the case of toxin-negative direct results.
Ethical disclosuresProtection of human and animal subjectsThe authors declare that no experiments were performed on humans or animals for this investigation.
Confidentiality of dataThe authors declare that they have followed the protocols of their work center on the publication of patient data.
Right to privacy and informed consentThe authors declare that no patient data appears in this article.
Conflict of interestThe authors declare that they have no conflicts of interest.
The authors acknowledge to MEDICA-TEC SRL and BIOARS SA from Ciudad Autónoma de Buenos Aires, Argentina, for kindly supply the commercially C. difficile toxin detection kits needed for the study.