In the field of genomic science and medicine in general, there are two methods of retrieving information from documents, namely: 1) through the combined use of associations determined by the Medical Subject Headings, and 2) by employing specific terminologies, such as those in folksonomies, alternative medical-genomic terms in use in the general language, or acronyms or apocopes from the genomics field. To some extent, many thinkers in matters of indexing hold that the combination of two methods may be the best approach. While few authors advocate for keeping the structure of controlled vocabularies, built up over many years of content interpretation, unchanged, there are numerous proposals for expanding the search horizons of thesauri, whether through social cataloging, algorithmic domain analyses that contrast indicators or the semantic web, using markers of meaningful semantic lexicons contained in digitized text.
Existen dos métodos de recuperación de información de documentos propios de la ciencia genómica y de medicina en general, a saber: uno está basado en el uso combinado de las relaciones determinadas por el Medical Subject Headings, mientras que el otro emplea las terminologías particulares, como pueden ser folksonomías, nombres alternativos de los términos médico-genómicos de uso en el lenguaje más general o los acrónimos o apocópes comunes en áreas como la genómica. Numerosos teóricos e indizadores consideran que la combinación de dos métodos puede funcionar mejor y es capaz de ofrecer mejoras significativas. Pese a que son pocos los autores que pugnan por no modificar la estructura de los vocabularios controlados, construidos a través de años de interpretación de contenidos, la multiplicidad de propuestas se reúnen bajo la tendencia de expandir el horizonte de búsqueda de los tesauros, ya sea con la catalogación social, el análisis de dominio realizado con algoritmos que contrastan indicadores o la web semántica, a través de la propuesta de marcado de unidades lexicales significativas en los textos digitalizados.
The Medical Subject Headings (mesh) is a body of terms put together by the National Library of Medicine (nlm) of the United States of America. Through the PubMed free access search engine, these lexical units are used to index and retrieve documents in the fields of biomedicine, genomic science and associated areas of knowledge from the citation and abstracts data bases. Both resources are also offered by nlm. PubMed offered a wide variety of notably efficient automated tools (PubMed Tools), including BioSample, Assembly and Genome and others. Genome, for example, organizes information from the area of genomics and includes sequences, maps, graphic representations of chromosomes and annotations by means of three main procedures. Each of these tools employs a distinct information retrieval method. In the case of Genome, thesauri structures have proven to be the most effective representation of information for the purpose of retrieval (Chute, 2005). Nonetheless, in recent years other methods and approaches have been proposed that seek to expand the possibilities of indexing and consultation of documents, though without abandoning the use of mesh (Bodenreider, Rindflesch and Burgun, 2002: 54).
Several of these proposals rely on the fact that most of the terms have several denominations. For example, phytomenadione is a synonym of vitamin K, and dihydroxyacetone phosphate is also know by the anagram dhap. It is also common to see the use of apocope, such as cocidioidosis for coccidioidomycosis. Moreover, they may be acceptable alternate spellings of concepts (Zweigenbaum and Grabar, 2004). While a search is generally perform using only a single terms, this method enriches the search criteria with derivations and alternate names for the concept sought. When there are a variety of names for a given concept, this does not mean that one is deemed correct at the expense of others. In the field of genomics, for example, synonymous terms are used in distinct contextual situations. A text retrieved from a public health science journal is not the same as that retrieved from a genomic map of cancer published in a highly specialized journal. Both documents are scientific in nature, but their respective outlooks are quite distinct, and they are targeted at different readership and satisfy different informational needs. Similarly, an academic discussion in an article on ascorbic acid (c6h8o6) is not the same thing as an informative blurb on vitamin c, even though the substance being discussed is the same chemical. Clearly, in such a case, the writers’ respective purposes are very different.
RETRIEVAL APPROACHES USING STRUCTURE OF A THESAURUSThe degree of specificity and the community are not the only criteria for distinguishing between the diverse uses of a term. bireme [Latin America and Caribbean Center of Health Information], which was first founded in 1967 as the Regional Library of Medicine with the support of the World Health Organization (who), developed the Health Science Descriptors (hsd) based on mesh, incorporating Spanish and Portuguese terms from the fields of homeopathy and sanitary oversight fields, as well as terms in English. This vocabulary supports the Virtual Health Library (vhl) and lilacs, which is the most important health index in Latin America and the Caribbean. There are communities, such as the Francophone, that do not choose to use medical headings and are more inclined to use names that diverge from those authorized by nlm. This happens in part because of the linguistic play of daily life and the adoption of the lexicon of the community. It is likely in such a case, that we would be speaking about folksonomies (Zweigenbaum et al., 2003). For example, the Catalogue et Index des Sites Médicaux de Langue Française (cismef) uses mesh and other vocabularies with metadata, which can exploit the rigor of the controlled vocabulary in conjunction with social cataloguing alternatives gathered from the community (Deacon, Smith and Tow, 2001). Some authors, such as Mary Rajathei David and Selvaraj Samuel, have proposed the Frequent Nearer Terms of the Domain (fntd) designed by Pubmed, as a method of retrieving information more efficiently. Such terms may or may not be authorized by nlm, and include derivations. The key criteria is that they are terms genuinely employed on a daily basis in the medical community (Rajathei David and Samuel, 2012: 20).
The editorial aim of cismef is to establish precise descriptions of documents largely on the basis of modified or improved of mesh. It constantly explores new descriptive approaches for retrieval of medical information (Kerdelhué, 2007). Some cismef contributors, working in the University Hospital of Rouen and led by Magaly Douyère, have attempted to adapt the broader, more general medical terminology used on the internet, instead of first resorting to scientific articles in the medline bibliographic base data. As already stated, cismef employs two standard tools to organize information: mesh and several subsets of Dublin Core metadata. The heterogeneous nature of online health information resources, however, led the cismef team to look for ways to improve mesh, first by designing a random algorithm assigning certain values to semantic links (Névéol et al., 2004), quite exhaustive but insufficient; and then by introducing the concepts of resource type and meta-terms. A resource type describes the nature of the document, not only the topic, as it happens with key words and mesh based qualifiers. A meta-term is often a broad term, such as the name of a discipline or medical treatment, offering semantic connections between mesh and the types of resources. cismef offers simple and advanced search options. The simple search requires the user to enter a single term or expression. This is complemented by a complete text search. The advanced search option performs complex searches with a combination of Boolean operators emplyed with meta-terms, key words, alternate names and resource types. This approach combines two tools to perform the search, i.e., mesh and the Dublin Core metadata format. As such, the documents are described jointly with the two tools. As such, documents are described with title, author or creator, topic, key words, description, editors, date, resource type, format, identifier and language (Darmoni et al., 2001: 167).
Bundschus and colleagues at the University of Munich and the company Siemens have opted for the term meta-information, which they use to complement the information, instead of the explicit distinction between the types of resources and the meta-terms used in France. The question is that by enriching medical information systems by including new documents while indexing with mesh terms, descriptions can be completed with this additional information: “This meta-information provides a rich source of knowledge that can be exploited in order to discover biomedical knowledge and data mining tasks” (Bundschus et al., 2008: 11). Bundschus adds: The term/concept model discovers new information from a set of biomedical texts, including the extraction of the structure of the concept of a hidden topic, using all of the mesh terms that concur within that subset […]. In contrast to standard topic models, in which the topics are represented exclusively by the most likely words, the topic-concept can be interpreted as a richer topic representations, especially by the link to mesh concepts. As such, this enriched topic representation provides important additional information as a terminological ontology (Bundschus et al., 2008: 18).
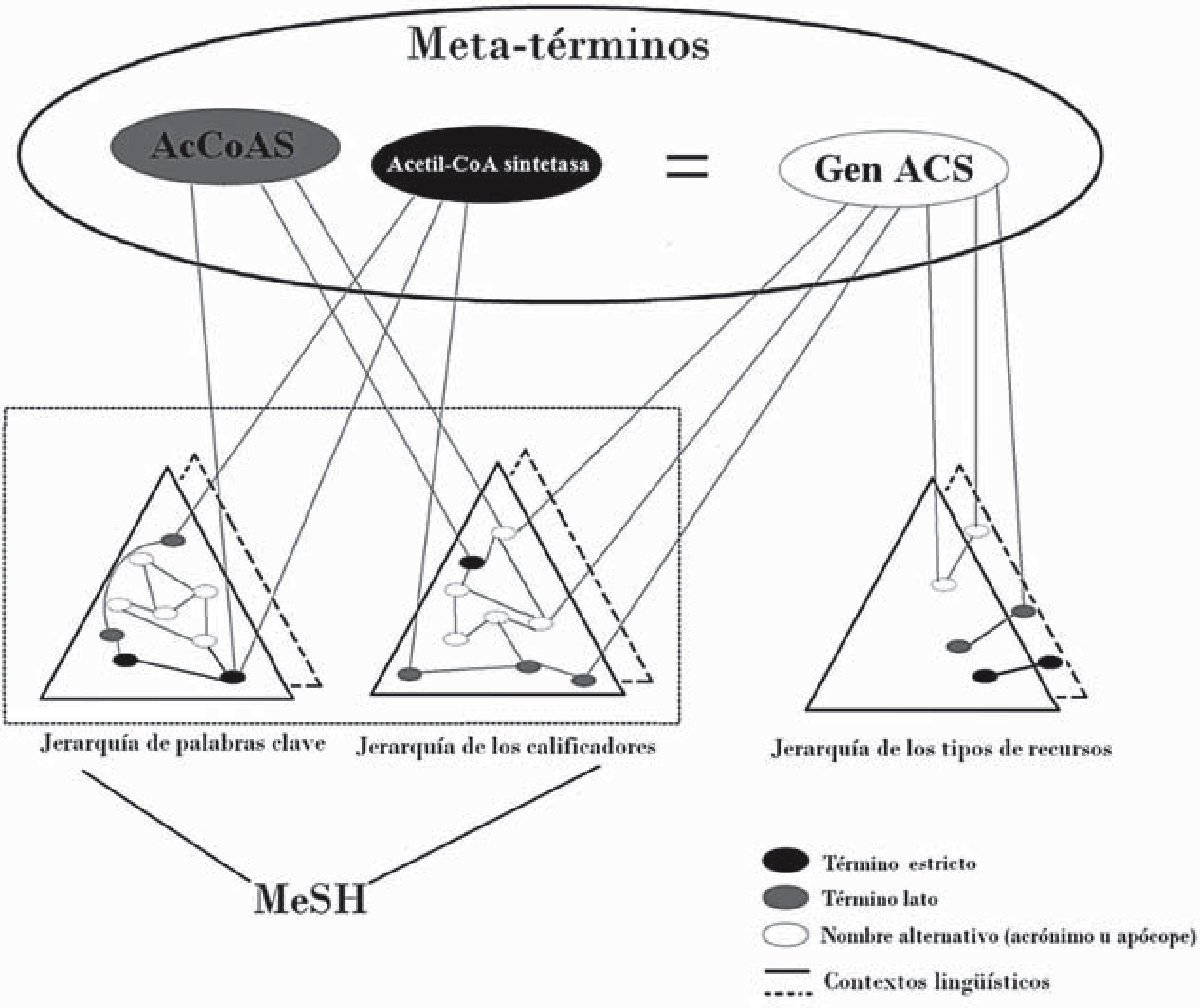
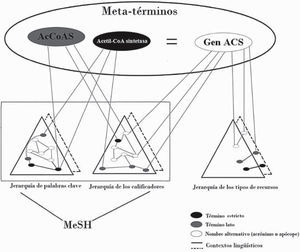
The Bundschus sciencometrics team has also explored applications such as extraction of statistical relationships between generic topics and mesh terms for the purpose of automated extraction of information (Leydesdorff, Rotolo and Rafols, 2012). In accord with proposals that do not circumscribe searches to authorized terms, two Spanish research projects show that gene accoas has nine alternate names in the biomedical literature, all of which are registered in the main human genome catalogue, the Online Mendelian Inheritance in Man (omim) as follows: cg9390, acetato-coenzima-A-ligasa, acetil-coa sintetasa, acetil coa sintasa, Acetil coa sintasa, acs, Acetil-coa synthasa, Acetil coa sintetasa, and best:-gh2840 (Galveza and Moya-Anegón, 2006: 345). On the basis of this example, the operation of the cismef proposal can be shown (Figure 1).
Exemplification of the functioning of CISMeF
Source: Douyère et al. (2004: 255)
cismef's efforts to renovate mesh is not new. In the early years of the twenty-first century, the Gesundheitsinformationsnetz Österreich (Austrian Health Information Network, gin), offered information to patients (identified as knowledge consumers), regarding not only preventive medicine, but also reliable medical information regarding illnesses, well-being and easily understood instructions for management of illness and access to information to better understand diagnoses. The system also provides specific data on the Austrian health system and organizations. Even though the description by gin were initially controlled by the mesh thesaurus, it was observed that users often did use scientific terms and expressions to understand their diagnoses. For this reason, they tried to match colloquial terminology to the more rigorous terminology using the vectorial method, in such a way that users were able to use the information system (Göbel et al., 2001: 242-244). Like the cismef, they used the informatics algorithm to perform automated searches, in this case the Floyd-Marshall algorithm.
Meanwhile, the Dutch researchers Radu Serban and Annette ten Teije (2009) favor the controlled vocabulary as an information representation and retrieval tool, believing that its structure should not be modified, nor should its specialized vocabulary be alternated with a more colloquial lexicon. In contrast, Edgar Meij and his group of archivists at the University of Amsterdam assert that the best method for retrieving information should be based on complex relationships of a controlled vocabulary. They hold that each descriptive record of a mesh term must be equal to a document on that term (and not that a term be equal to a document or that it should retrieve several documents). With this idea, it would be inadmissible to discard an immense variety of alternate names for a term that are useful for the expansion of the methods of information retrieval and are more effective representation of a document than that supplied by mesh alone (Meij et al., 2005; Nelson, Johnson and Humphreys, 2001: 177).
In the case of a Swiss epistemic community, the particular terminology of a group of researchers is used, specifically that of the Swiss-Prot Group (a question that often concerns such communities). This research group alternates the use of mesh with their particular terminology in order to make search more reliable (Mottaz, 2006: 18). More than ten years ago, Dieuwke Brand-de Heer adduced: […] medline certainly does not cover “the totality” of medical literature. Other data bases contain additional information; for example, Excerpta Medica also covers medical topics. Moreover, for some fields these alternative data bases behave better than medline, for example in pharmacology. There is also biosis previas, which contains additional relevant information for doctors not included in medline (2001: 112).
The Slovenian specialist in medical literature Tomaz Bartol holds that the use of broader terms is useful, because it improves the retrieval of relevant documents. He recently performed a study on the information on herbal medicine, arguing that: In our study, we have placed special emphasis on the importance of the question of co-occurrence of different terms, especially descriptors, in the same document. This type of research generally entails descriptors based on a dictionary of synonyms, such as mesh. The alternative terms and names hold meaning only in the “contexts of their use.” The traditional classification systems, however, are often resistant to context. The indexation terms in thesauri are generally based on vast, rigid structures and predefined hierarchies, which do not always perform effective retrievals within a given topic area (Bartol, 2012: 286).
In accord with Meij, Bartol asserts the need to expand the horizons of searches by using controlled vocabularies and enriching their structure. Bartol provides the example of the term dittany, which in many data bases is conflated with the term salvia. According to Meiji and his team, using the term dittany would not retrieve the same documents that would otherwise be retrieved by the term salvia, despite the fact that both terms refer to the same plant. This is because these denominations denote distinct contexts and will bring back different documents (“each term for each document”). Bartol points out that it would be a mistake to tie both terms together in the description in order to ensure retrieval of the same document from a data base using either search term. Each search term should represent a distinct information need.
UNIFICATION OF MEDICAL LANGUAGELittle by little, the use of alternative names has influenced the structure of mesh itself, and more so now that records contain not only medical headings as descriptions, but also alternative names. These description include notes on the scope of nomenclature, acronyms and references to names used previously. For several years, the National Library of Medicine has a linguistic tool working in addition to the medical topic headings. This tool is the Meta-thesaurus of the Unified Medical Language System (umls), in which other medical information systems also collaborate. As technology progresses, it is used to create new ways to index for the purpose of information retrieval. The Macro-thesaurus aims initially to be an ontology that integrates the knowledge of diverse thesauri and other sources. This done not in order to expand the search, but to specify the retrieval of information (Humphreys and Schuyler, 1993).
In this sense, Hassan, Htroy and Palombi (2010) propose two main approaches for representing the medical knowledge:
- •
Image based focus: classic atlas, informatics atlas and probabilistic atlas. These atlases provide a model for some organs and the labeling of these organs is often manual.
- •
Ontological based focus. An ontology is, by definition, a formal representation of a subset of concepts within a domain, in addition to the relationships among these concepts.
“An ontology is a formal specification of shared conceptualization.” This definition was coined in 1998 by Studer, Benjamin and Fensel. Pastor Sánchez has taken it up again and provides the following exposition: The term conceptualization refers to an abstract model of concrete reality that is obtained by identifying the concepts relevant to the same. By explicit, we mean that the type of concept used and the restrictions of its use are explicitly defined. Formal refers to the fact that the ontology should be legible for the computer; and shared reflects the notion that an ontology captures knowledge that is not object of a single individual, but rather accepted by a group in a consensual way (2011: 20).
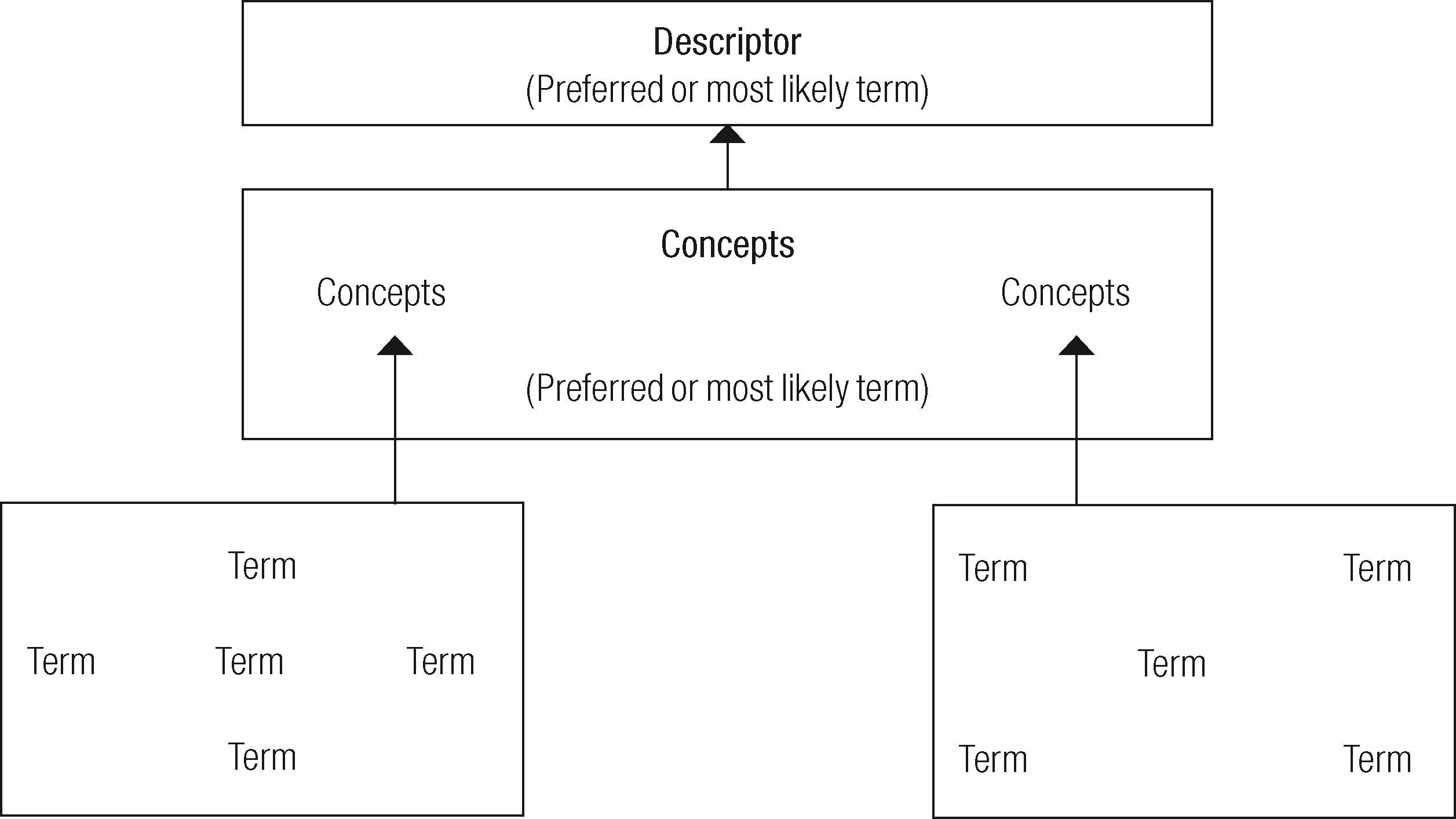
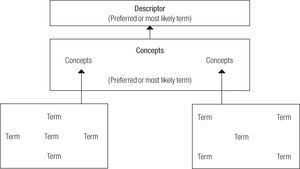
The Meta-thesaurus is only one of the tool of umls, which is also integrated by other tools such as Semantic Network and specialist Lexicon. The umls project has developed slowly. It attempts to combine three tools to achieve effective retrieval of information. The Meta-thesaurus is in charge of the concepts; semantic Network of the categories and relationships, and specialist Lexicon is in charge of the resources and tools (Kostoff et al., 2004: 518). Meta-thesaurus began in 1988 and is constituted on the basis of automated versions of diverse thesauri and heading lists (in diverse languages other than English, including Spanish, French. Dutch, Italian, Japanese and Portuguese), codes and lists of controlled terms used in patient care, such as gin, public health statistics and the indexation of biomedical literature. The terms in the Meta-thesaurus are organized by meaning, and they are assigned a unique concept identifier (with several associated lexical identifiers). All the original data of the source vocabulary, the definitions or written variants are organized. The mesh has been limited by the delay in the adoption of new terminology. Meta-thesaurus, moreover, does not always incorporate the newest topics in a timely way. For this reason after 2004, the use of metadata by Meta-thesaurus produced a weighty change in the way documents are managed and nml formats (Figure 2).
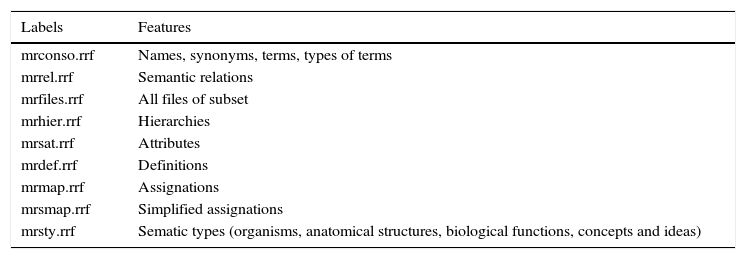
In order to allow for the complex description that includes acronyms and abbreviations indexed in the Systematized Nomenclature of Medicine, umls developed the Rich Release Format (rrf) (Chute, 2005: 176). For complete retrieval of information of both mesh terms and alternative names, better search strategies must be incorporated. Nonetheless, Meta-thesaurus has the merit of using metadata to provide a greater search scope, which with headings is insufficient. It has nearly forty labels, the most representative of which are the following table.
Main labels in Rich Release Format (RRF).
| Labels | Features |
|---|---|
| mrconso.rrf | Names, synonyms, terms, types of terms |
| mrrel.rrf | Semantic relations |
| mrfiles.rrf | All files of subset |
| mrhier.rrf | Hierarchies |
| mrsat.rrf | Attributes |
| mrdef.rrf | Definitions |
| mrmap.rrf | Assignations |
| mrsmap.rrf | Simplified assignations |
| mrsty.rrf | Sematic types (organisms, anatomical structures, biological functions, concepts and ideas) |
These functions enrich the Meta-thesaurus. This is only one of the three UMLS tools. (Figure 3).
Operation of umls Meta-thesaurus search on the basis of metadata records and the relationships established between terms, concepts and descriptors.
Source: Mottaz, 2006: 8
From the perspective of informatics, Christopher Chute adds: “Previously, the umls process of formatting performed information transfer ‘with losses.’ The modern vision of umls is to become a definitive source and format for publication of the main biomedical terminologies, which is a significant advance” (2005: 176-177). This means that the pretension of unifying language by uml is not only a matter of terms, but also a question of improving the computational systems. umls have tried to establish an information exchange format for the medical area that little by little contributes to this end. Nonetheless, the most advanced projects is still the cismef: In cismef, resources are described using a set of metadata on the basis of a structured terminology that “encapsulates” the French language version of the mesh thesaurus. Now the objective is to migrate the cismef terminology and, thereby, mesh to a formal ontology in order to obtain a more powerful search tool (Soualmia, Golbreich and Darmoni, 2001: 1).
Currently, the particular terminology of cismef has been “formalized” on the Web Ontology Language (owl), in its dl version, in contrast to the ontologized thesauri that are in the owl-Full version.
CONCLUSIONSDiverse cases across the international scenario point toward the need to expand the horizons of representation and retrieval of information. The structure of the controlled vocabularies should be complemented by other methods. Despite the general belief that the use of layman language brings back a list with an enormous number of results, the reality is that, in the area of health, the terminology can be very specific when discovering a document, and terms in the strict sense can very likely be retrieved. Finally, sooner or later, the thesaurus shall have be integrated fully with the semantic web, perhaps as onto-thesauri (if one wishes to view thesauri as ontologies). In genomic sciences and health, the combination of the semantic relationships of a thesaurus, a terminology alternative and the metadata and search engines would create a tool with unimaginable potential. This is an important endeavor for the field of Library Science research.