Raman spectroscopy is a vibrational spectroscopic technique for assessing molecular motion and fingerprinting species, which can be used for the identification of molecules in living cells. We have obtained Raman spectra from HEK293T cells expressing a single transgene in order to analyze whether this technique could be of interest as a tool for identifying subtle changes in gene expression. Spectrum obtained from cells expressing the transgene exhibit features indicating that active vibrational modes related to phenylalanine residues are dominant. Similarly, CO stretching mode of a functional carboxylic acid group is absent in transfected cells. In addition to these significant changes, oscillatory strengths of several vibrational modes are altered. The present analysis suggests that Raman spectrum could be a tool to identify changes due to the expression of a single gene.
La espectroscopía Raman es una técnica espectroscópica vibracional que permite medir movimientos moleculares y obtener perfiles moleculares, y que se puede utilizar para identificar moléculas en células vivas. Nosotros hemos obtenido espectros en células HEK293T que expresan un transgén, al objeto de comprobar si esta técnica podría ser de interés como herramienta para identificar pequeños cambios en la expresión génica. El espectro obtenido en células transfectadas es indicativo de que los modos vibracionales del aminoácido Fenilalanina son predominantes. De manera similar, el modo vibracional de tipo CO característico de los grupos carboxílicos está ausente en células transfectadas. Aparte de estos cambios, también están alteradas las fuerzas oscilatorias de otros modos vibracionales. El presente análisis sugiere que los espectros Raman pueden constituir una herramienta de utilidad para identificar cambios debidos a la expresión de genes específicos.
Raman spectroscopy is a powerful technique for optical diagnostic.1,2 Recently, Raman spectroscopy together with Raman imaging have been applied for the early detection of cancer.3 This is because it provides fundamental information about vibrational modes of a given system. Eigenvalues4 as well as the oscillatory strength of vibrational modes are very sensitive to the nature of chemical bonds and their alterations due to perturbations originated from the environment of a specific bond as well as the attachment of functional groups such as NH2, CO, etc.5,6 Addition or substitution of chemical groups modify the charge distribution and alters the dipole strength of the vibrating unit.7 All these parameters could be profoundly modified with variations in gene expression. Therefore, Raman spectroscopic techniques could be a unique and direct approach for evaluating changes of gene expression at a molecular level. Therefore, the purpose of the present work is to examine Raman spectral variation upon expression of a single transgene.
Modifications in the expression of a single gene is seldom accompanied by morphological changes, and is thus identified by means of monoclonal antibodies specifically recognizing gene products, or by the simultaneous expression of fluorescently active genes such as GFP (Green Fluorescent Protein). Currently there are no means by which we could identify expression of single genes unless we perform additional labeling and, therefore, Raman spectroscopy is found to be an adequate tool to obtaining this type of information.
In the present work, we analyze in an easily transfectable cell line such as HEK293T (Human Embryonic Kidney cells) changes induced in Raman spectra by the expression of a single transgene.
Matherial and methodsPlasmids, cell culture and transfectionsHEK293T cells (Invitrogen), were seeded at 2–5×104 viable cells/cm2, maintained at 37°C in 5% CO2 in Dulbecco's modified Eagle's medium (DMEM; GIBCO) containing 10% fetal bovine serum, 0.1mM MEM Non-Essential Amino Acids (NEAA), 2mM L-glutamine and antibiotics (130U/ml penicillin and 130μg/ml streptomycin) and subcultured when reaching 80% confluency. One day prior to transfection 2×104 cells were seeded in 500μl growth medium without antibiotics in a 24-well plate. After 24h, cells were transfected with the pEGFP expression plasmid containing the cDNA coding for GFP, using Lipofectamine™ 2000 reagent (Invitrogen). Twenty-four hours after transfection, cells were subcultured in fresh growth media and seeded onto cover slips coated with poly-d-Lysine to prevent detachment during in vivo Raman analysis.
Raman spectroscopic analysis of HEK293T cellsFor Raman analysis a cover slip with bound HEK293T cells was transferred to a cuvette containing PBS (NaCl 0.138M; KCl: 0.0027M; pH 7.4 at 25°C), a transparent buffer, in order to minimize interference. Raman spectra was obtained at room temperature, from the central portion of the cells by a Renishaw Raman Microscope System RM2000 equipped with a diode laser of 785nm, a Leica microscope and an electrically refrigerated CCD camera as a detector. The spectra were obtained with a 100× magnification. During acquisition, the power of the incident radiation was maintained at a low (2.0mW) level to keep cells alive. Spectral resolution was set at 2cm−1. The area from which Raman radiation was collected was of the order of 1μm2.
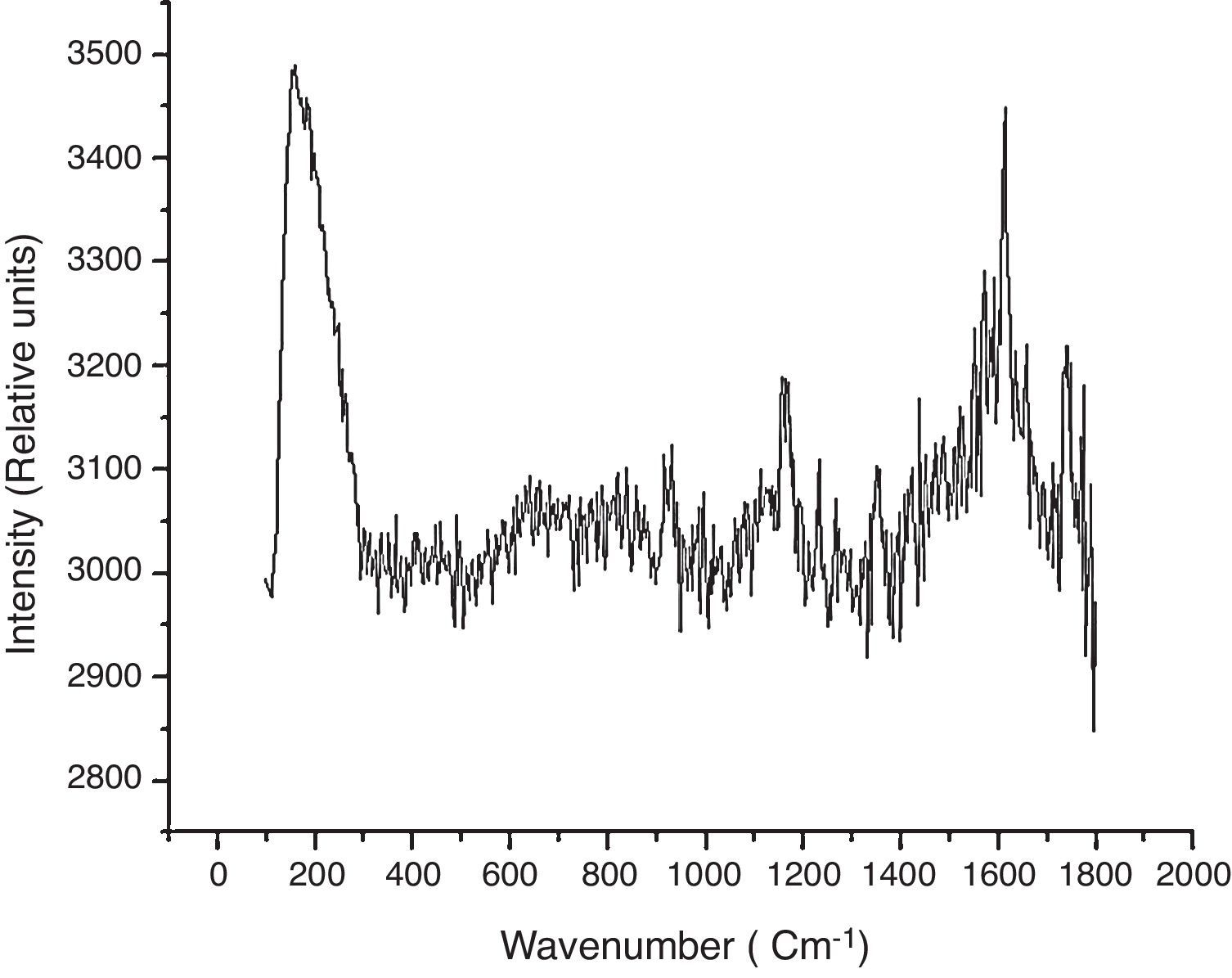
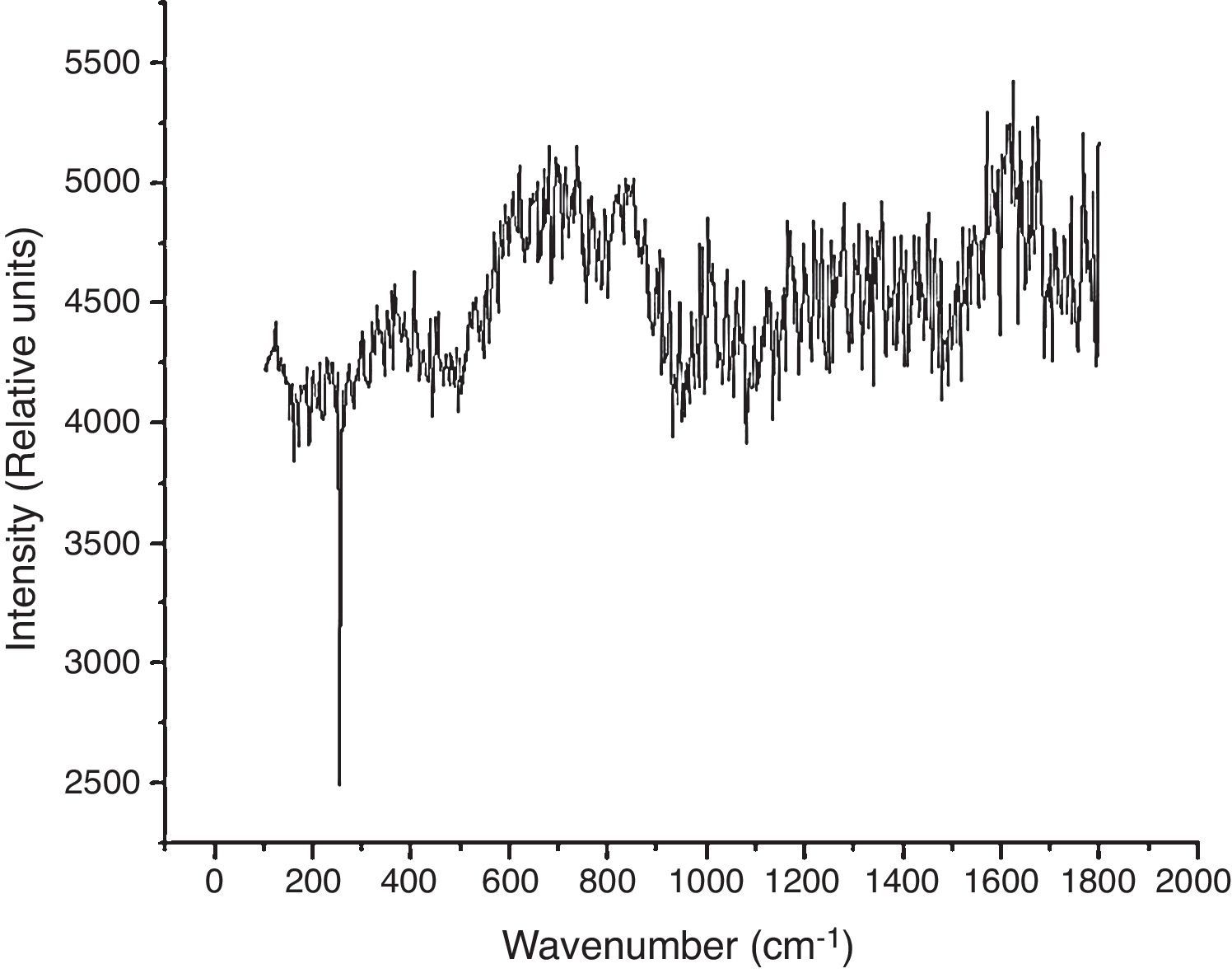
Results and discussionComparison between spectra obtained in non-transfected and GFP-transfected HEK293T cells clearly shows that the presence of GFP substantially changed vibrational mode patterns. The peak located at 1603cm−1 (Fig. 1), along with its broad structure, is changed completely and instead a narrow structure comes out. Similarly, the peak located at 1175cm−1 disappears, while a weak broad shoulder remains. Meanwhile, the structure between 600 and 800cm−1 is enhanced. Thus, the expression of a single protein completely changes the vibrational modes (including oscillatory strengths) suggesing that Raman spectra could be a direct approach for the identification of subtle changes in gene expression.
To understand the origin for this behavior, it is necessary to examine the source for these peaks. The structure located between 600 and 800cm−1 could be associated with the mode of ring breathing6,8 of Phenylalanine [F] or alternatively to the carbon–sulfur stretching mode that is also present.8 We favor the implication of Phenylalanine based on the abundance of the Phenylalanine ring as compared with carbon–sulfur bonds. Moreover, earlier reported studies9 show that Phenylalanine has vibrational modes located at 617, 754 and 822cm−1 and a careful evaluation of the broad peak, suggests that it consists of three unresolved peaks located in the region close to 615, 750 and 820cm−1. In addition there is another indication of the domination of Phenylalanine vibrational modes such as a small, broad but clear peak located at 410cm−1. In general, in this region no peak is expected when one examines Raman spectra of amino acids or proteins. Only in a few selective cases, such peak is observed, particularly when a CH2 group is attached to an aromatic group10 as is the case for Phenylalanine. Thus on the background of previous considerations, this peak could be attributed to Phenylalanine vibrational mode.
A comparative analysis of the spectra from untransfected and GFP-transfected cells, reveals that the vibrational peak located at 1734cm−1 is absent in the later (Fig. 2). It is known that in carboxylic acids, the functional group corresponding to CO stretching mode8,10 is located at 1730cm−1. The close agreement between the experimentally observed value and the reported one, suggests that this peak is originated from CO stretching vibrational mode. This mode is present in untransfected cells, but it is virtually absent in GFP-transfected cells. The reason for this change is not a substantial change in the concentration of carboxylic, but a modification of its environment leading to a decrease in the oscillatory strength corresponding to this vibrational mode.
Another notable change is observed in the region 1400–1700cm−1. The spectrum obtained from normal cells show a relatively sharp peak (located at 1619cm−1) while in GFP-transfected cells, this dominating structure is absent and instead a broad band without any structure is observed. In addition to this, the other complexity arises from the simultaneous presence of Raman lines due to other amino acids such as Proline [P], Methionine [M], Leucine [L] and Glycine [G] in the same region, with the superposition of these structures making the identification even a more difficult task. Thus the origin of this peak remains to be elucidated.
A peak with a broad shoulder is observed at 1165cm−1 in non-transfected cells, but it is completely absent in GFP-transfected cells. A likely origin for this peak is CH stretching mode when C atom has two or three substitution groups at the other bond sites. In the present case the most probable group could be Valine [V], which has a CH group with the central carbon atom attached to two groups of CH3 units. The reported range of frequencies is quite large and covers the range from approximately 1000–1200cm−1. In the present investigation, the structure is observed exactly in the same range and therefore, this peak could be attributed to CH vibration. Infrared studies show that the vibration of CH mode is slightly out of the plane,10 thus even in non-transfected cells there is also some degree of distortion which forces the hydrogen atom out of the plane which is concordant with complex structures.
The NH2 group constitutes the most important vibrational feature of amino acids, giving the presence of a rather broad band9 located in the range 1585 to 1685cm−1. This is very often called as a deformation of the amino-acid bond where the NH stretching mode is rather out of the plane and over all the amino acid is deformed from the planer point of view. This structure is observed in GFP-trasfected cells (Fig. 2). This band has low intensity and very often is mixed in the noise level. However, because of the improved experimental technique and the good detector (CCD operated at low temperature), the structure is visible even though it is mixed with the noise. This noisy structure does not allow for a detailed analysis but suggests that some amino acids such as Glutamine [Q], Glutamic [E] or Asparagine [N] are present, dominating the vibrational spectra in this region.
ConclusionsRaman spectral analysis was carried out to examine variations induced by the expression of a single gene in HEK293T cells. Here we show that changes in the vibrational modes are substantial suggesting that Raman analysis could be adequate for gene expression analysis.
Ethical disclosuresProtection of human and animal subjects. The authors declare that no experiments were performed on humans or animals for this investigation.
Confidentiality of Data. The authors declare that no patient data appears in this article.
Right to privacy and informed consent. The authors have obtained the informed consent of the patients and /or subjects mentioned in the article. The author for correspondence is in possession of this document.
Conflict of interestThe authors declare no conflict of interest.
This work was supported by the Ministry of Science and Innovation (MICINN) (Spain) [grant number SAF2009-09449], by Consejeria de Salud (CS) [grant number PI-0007/2007] and by Consejeria de Economia, Innovacion y Ciencia, Junta de Andalucia Spain [grant number P08-CTS-04348] to F.G.-C.