The urgent need for operational research evaluating test performance in a real-world setting has been highlighted. The purpose of this study was therefore to evaluate the performance of MTBDRplus assay.
MaterialsAccording to the reference method, of the 155 clinical specimens with valid results, 147 were susceptible to rifampicin (RIF) and isoniazid (INH), with 4 being multi-drug resistant (MDR) and 4 with isolated resistance to isoniazid (INH).
ResultsThe results of the MTBDRplus assay were 100% concordant for the MDR and mono-resistant to INH specimens. However, the MTBDRplus assay showed a resistance pattern to RIF in one specimen which was classified as susceptible by the reference method. The majority of the specimens (118/75.6%) were also tested using the MTBDRplus method after culture on Lowenstein-Jensen media, showing 100% agreement with the results of the test directly from the specimens. An MTBDRplus test result was available within an average of 8 days.
ConclusionsOverall, MTBDR results showed excellent results when compared with the reference method and achieved a significant time-reduction.
Es importante evaluar el desarrollo de los ensayos moleculares en la práctica diaria del laboratorio de microbiología.
MaterialesSe incluyeron 155 muestras clínicas con resultado válido. De acuerdo con el método de referencia, 147 fueron sensibles a INH y RIF, 4 MDR y 4 presentaron resistencia aislada a isoniazida.
ResultadosLos resultados del ensayo Genotype MTBDRplus fueron concordantes 100% en la muestras MDR y con resistencia aislada a isoniazida. Sin embargo, el ensayo Genotype MTBDRplus demostró un patrón de resistencia a RIF en una muestra que fue sensible por el método de referencia. En 118 muestras (75.6%) también se realizó el ensayo Genotype MTBDRplus sobre la cepa obtenida tras cultivo en medio Lowenstein-Jensen, mostrando un 100% de concordancia con los resultados obtenidos por el test directamente en muestra clínica. De media, los resultados del ensayo Genotype MTBDRplus estuvieron disponibles en 8 días.
ConclusionesEn conjunto, el ensayo Genotype MTBDRplus mostró resultados excelentes cuando se comparó con el sistema de referencia y consiguió una reducción significativa en el tiempo de emisión de resultados.
The rapid detection of Multidrug resistant tuberculosis (MDR-TB) ensures treatment with appropriate drugs, thereby reducing morbidity, mortality, economic costs, the further transmission of infection and the emergence of extremely drug resistant strains.1 For these reasons, it is important for clinicians to receive rapid susceptibility results from the laboratory. By using molecular biological methods, such as line probe assays, to detect mutations that are associated with drug resistance against rifampicin (RIF) and isoniazid (INH), preliminary susceptibility results can be reported within a few days after receiving the sample. While tests such as the Genotype MTBDRplus assay offer great promise to improving MDR-TB detection and care, the urgent need for operational research evaluating test performance in a real-world setting has been highlighted.2
Thus, although the GenoType® MTBDRplus version 1.0 assay has been studied by several authors, this publication is the first of its kind in Almería province, where a moderate incidence of TB has been described (average incidence rate: 26.6 cases/100,000 inhabitants).3 The purpose of this study was therefore to evaluate the performance of MTBDRplus assay for the rapid detection of Mycobacterium tuberculosis and mutations causing RIF and high or low level INH resistance in an attempt to compare with the results obtained by our reference susceptibility method. The assay was performed directly in smear-positive pulmonary specimens and also in some of the M. tuberculosis isolates grown from the sub-culture from liquid media to Lowenstein-Jensen with objective of comparison. The MTBDRplus assay is compared to drug susceptibility testing performed by automated liquid culture system (BACTEC MGIT 960, BD, Sparks, MD, USA).
Materials and methodsClinical specimensTwo hundred and fifty-two respiratory samples (225 sputa, 22 bronchial aspirates, 1 bronchoalveolar lavage and 4 gastric aspirates) from 247 patients (for each patient, only one specimen with the highest number of AFB on stain was taken into account) were prospectively selected from the samples collected between December 2009 and December 2011 during the clinical routine at the Torrecárdenas Hospital (Almería, Spain). Before they were tested, all samples were first digested and decontaminated by using the Kubica N-acetyl-l-cysteine NaOH method.4 Later, auramine-rhodamine acid-fast staining was performed on the concentrated sediment as we have already described.5 During the study period, samples-selection criteria were AFB on stain but only diagnostic specimens were included. There are more samples tested than patients included because in case of a MTBDRplus invalid result with the first specimen, another one was tested. An aliquot of the decontaminated specimens was cultured on BacT/Alert liquid medium (BioMérieux, Marcy l’Etoile, France) and Lowenstein-Jensen solid agar. The identification of M. tuberculosis complex was confirmed in cultures by the AccuprobeMTC assay (Gen-Probe, Inc., San Diego, CA).
Drug susceptibilityTesting for susceptibility to INH, RIF, pyrazinamide, ethambutol, and streptomycin was performed with the automated BACTEC MGIT 960 by using critical concentrations of 0.1μg/ml for INH and 2μg/ml for RIF at a level III Regional Reference Laboratory (Hospital Costa del Sol, Marbella, Spain).
GenoType MTBDRplus assayThe Genotype MTBDRplus assay (Hain Lifescience, Nehren, Germany) was performed directly from the 252 decontaminated smear positive respiratory specimens, before the culture results were obtained. Identification culture results found MTC in 177 (70.2%) specimens. Cultured sample on Lowenstein-Jensen material was available in 118 samples out of 177 (66.7%), in which the molecular assay was also performed. All tests were performed according to the manufacturer's instruction.
Genotypic characterizationOne of the specimens showing disagreement between the molecular and the reference method was sequenced at a Reference Laboratory (Hospital Gregorio Marañon, Madrid, Spain).
Data analysisThe turnaround time of results from the molecular and the reference methods was compared. The data were analyzed by using SPSS software (version 15.0). Comparisons were done by chi-square analysis. A P value of ≤0.05 was considered significant.
ResultsMTBDRplus assay results for the clinical specimensValid results were obtained for one hundred and fifty-five (155/252, 61.5%) specimens. Among the 252 specimens, identification culture results found MTC in 177 (70.2%). In 75 specimens, culture was negative (43; 17%), contaminated (7; 2.7%) or a NTM was isolated (25; 9.9%). Among 177 molecular tests performed in specimens yielding MTC in culture, 12.4% (22/177) were not interpretable due to: absence of MTC specific band (7 paucibacillar specimens, 4%), lack of any locus control band (14 specimens, 8%) and absence of more than one rpoB wild type band (1 specimen, 0.4%). None of the negative, contaminated or NTM cultured specimens showed an interpretable band pattern on the molecular test as all of them lacked the specific MTC band.
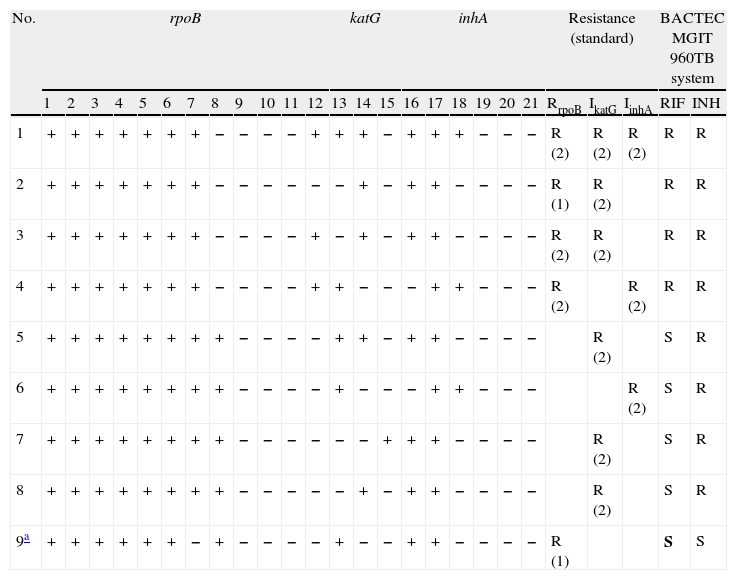
Among the 155 clinical specimens with valid results, 147 were fully susceptible by the reference method, being 4 MDR and 4 mono-resistant to INH. The specificity of the assay for the detection of resistance to RIF, INH and MDR was 99.3%, 100% and 100% respectively. The sensitivity for the detection of resistance to RIF, INH and MDR was 99.3%, 100% and 100%, respectively. Thus, the MTBDRplus assay showed a RIF resistance pattern in one specimen which was classified as susceptible by the reference method. In this specimen, the mutations in rpoB identified by the assay were the lack of a band for the WT7 probe (sequence showed CAC>526>AAC) (Table 1, specimen 9). In two out of the five cases of RIF resistance detected by the molecular method, the criteria was the absence of a single wild-type signal corresponding to mutations at codons 526 and 530 (Table 1). Only in one case, this criteria of the molecular method was in agreement with the reference assay (specimen 2). In the other three cases, the appearance of a mutant probe band was the criterion for the RIF resistance (in all cases due to a serine-to-leucine amino acid substitution at codon 531 [S531L]). All cases of INH resistance were caused by the appearance of a mutant probe band, predominantly, in katG gene (6 cases versus 3 in inhA), showing high level resistance to INH (Table 1). katG most commonly associated mutation was S315T1. One patient (specimen 1) showed wild-type and mutant rpoB, katG and inhA hybridization signals and resistance to INH defined by mutations in both katG and inhA.
MTBDRplus band pattern in clinical specimens showing resistance to INH, RIF or both.
| No. | rpoB | katG | inhA | Resistance (standard) | BACTEC MGIT 960TB system | |||||||||||||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | RrpoB | IkatG | IinhA | RIF | INH | |
| 1 | + | + | + | + | + | + | + | − | − | − | − | + | + | + | − | + | + | + | − | − | − | R (2) | R (2) | R (2) | R | R |
| 2 | + | + | + | + | + | + | + | − | − | − | − | − | − | + | − | + | + | − | − | − | − | R (1) | R (2) | R | R | |
| 3 | + | + | + | + | + | + | + | − | − | − | − | + | − | + | − | + | + | − | − | − | − | R (2) | R (2) | R | R | |
| 4 | + | + | + | + | + | + | + | − | − | − | − | + | + | − | − | − | + | + | − | − | − | R (2) | R (2) | R | R | |
| 5 | + | + | + | + | + | + | + | + | − | − | − | − | + | + | − | + | + | − | − | − | − | R (2) | S | R | ||
| 6 | + | + | + | + | + | + | + | + | − | − | − | − | + | − | − | − | + | + | − | − | − | R (2) | S | R | ||
| 7 | + | + | + | + | + | + | + | + | − | − | − | − | − | − | + | + | + | − | − | − | − | R (2) | S | R | ||
| 8 | + | + | + | + | + | + | + | + | − | − | − | − | − | + | − | + | + | − | − | − | − | R (2) | S | R | ||
| 9a | + | + | + | + | + | + | − | + | − | − | − | − | + | − | − | + | + | − | − | − | − | R (1) | S | S | ||
1 WT1 (506–509), 2 WT2 (510–513), 3 WT3 (513–517), 4 WT4 (516–519), 5 WT5 (518–522), 6 WT6 (521–525), 7 WT7 (526–529), 8 WT8 (530–533), 9 MUT1 (D516V), 10 MUT2A (H526Y), 11 MUT2B (H526D), 12 MUT3 (S531L), 13 WT (315), 14 MUT1 (S315T1), 15 MUT2 (S315T2), 16 WT1 (−15/−16), 17 WT2 (−8), 18 MUT1 (C15T), 19 MUT2 (A16G), 20 MUT3A (T8C), 21 MUT3B (T8A).
Standard 1: lack of wild type band. Standard 2: presence of mutant hybridization signal.
Among the 155 clinical specimens tested directly, 118 (75.6%) were also tested by the MTBDRplus after culture on Lowenstein-Jensen media, showing 100% agreement with the results of the test directly from the specimens. One of the specimens showing disagreement in RIF resistance with the reference method also showed lack of the WT7 band when the assay was performed on the clinical strain.
Time to resultsTime to detection of M. tuberculosis and drug resistance was significantly shorter for the MTBDRplus assay compared to the conventional method. On average, an MTBDRplus test result was available by 8 days while results from the reference laboratory were not available until 41.5 days.
DiscussionThe performance of MTBDRplus assay directly from smear-positive sputum correlated very highly with BACTEC MGIT 960 results. As previously reported, the proportion of invalid results was correlated with smear status, with much higher failure rates in very low smear-positive specimens6.
In this study rifampicin resistance was most commonly associated with mutation in the region of rpoB 530–533, mostly S531L mutation, which has already been described7. This mutation was more frequently found in MDR strains than in rifampicin monoresistant strains. Isoniazid resistance was most commonly associated with katG S315T1 mutation.
One RIF-susceptible strain in this study revealed mutations in the selected rpoB loci. In this case, sequence showed CAC>526>AAC, however this mutation did not translate into phenotypic resistance. This absence of rpoB wild type bands not translated into RIF resistance has previously been described.8
This study is the first of its kind to compare results of MTBDRplus assay first directly on the clinical specimen and afterwards on the strain obtained from the clinical specimen’ culture on Lowenstein-Jensen agar. Assay results were 100% concordant.
One of the limitations of our study is the low prevalence of MDR isolates in our setting. For this reason, it is difficult to draw conclusions, except that of careful interpretation of the absence of rpoB wild type bands. Overall, MTBDR results showed excellent results when compared with the reference method and achieved a significant time-reduction, which might have important financial and clinical savings as a consequence. However, because only the most frequent mutations are detected, the conventional first line susceptibility testing is still needed.
Conflicts of interestNone of the authors have any conflicts of interest.
We are grateful to Aurora Cejudo, Natalia Montiel Quezel-Guerraz, Alejandro Barbancho, Jose Ramón Maldonado, Armando Reyes Bertos, Pablo Sánchez Villegas, M. Luisa Sánchez, Magdalena Bonillo, Silvia Vallejo, M. Cruz Rogado, Francisca Escabias, Manuel Alfredo Martínez, M. Dolores Navarro, Francisco Laynez and Isabel Cabeza, all of them members of the Indal TB Group for their contribution to this study. This study was partially supported by the Junta de Andalucía (PI-0444/2008 and PI-0306/2009) and the SEPAR (Spanish Society of Pneumology and Thoracic Surgery) (763/2009).