Legionella is an important cause of both community-acquired and nosocomial pneumonia. Human infection has two clinical entities: Pontiac Fever (a self-limited flu-like illness) and Legionnaires’ Disease (a severe multisystem disease involving pneumonia).1 The genus Legionella (L) includes nearly 60 species with a considerable number of serogroups (J.P. Euzéby: http://www.bacterio.cict.fr/). Of all species of Legionella, Legionella pneumophila (LPN) accounts for >90% of human infections and serogroup 1 for 70–80% of these. Legionellosis can also be caused by LPN serogroups 2, 3, 4, 5 or 6 and other L species (e.g., L. micdadei and L. longbeachae).2 Isolation by culture is still of major importance in environmental analysis and clinical diagnosis and is essential for epidemiological studies and to characterize the isolate causing an outbreak.1,3 The objective of this study was to evaluate a novel immunochromatographic test (ICT) designed for the identification of Legionella colonies from bacterial culture. The test includes two strips: one for L identification and another one with two test lines for LPN serogroups 2–15 and L. pneumophila serogroup 1 (LPN1) identification. Legionella identification is based on a combination of monoclonal antibodies (mAb) (developed at Vircell, S.L., Granada, Spain), adsorbed on both the colloidal gold and the membrane, targeting specific constituents of the bacterial cell wall: the peptidoglycan-associated lipoprotein (PAL) for L, the major outer membrane protein (MOMP) for LPN and the lipopolysaccharide (LPS) for LPN1. PAL was selected to detect genus because it is a constant and highly conserved antigen of the Legionella membrane.4 MOMP is useful to identify LPN strains because it is present in the membrane of LPN serogroups 1–15 but not in that of non-pneumophila Legionella.5 Finally, LPS is a highly immunogenic cell surface component that offers serogroup/mAb subgroup specificity. Most of the clinical isolates, especially those associated with outbreaks, contained a specific epitope for mAb 2 and mAb 3/1 from the Dresden Panel.6,7 Of these, mAb 3/1 reacts with the 8-O-acetyl group from legionaminic acid which is a key component of LPN1 LPS,8 confirming that LPS is a suitable antigen to identify serogroup 1 of LPN.
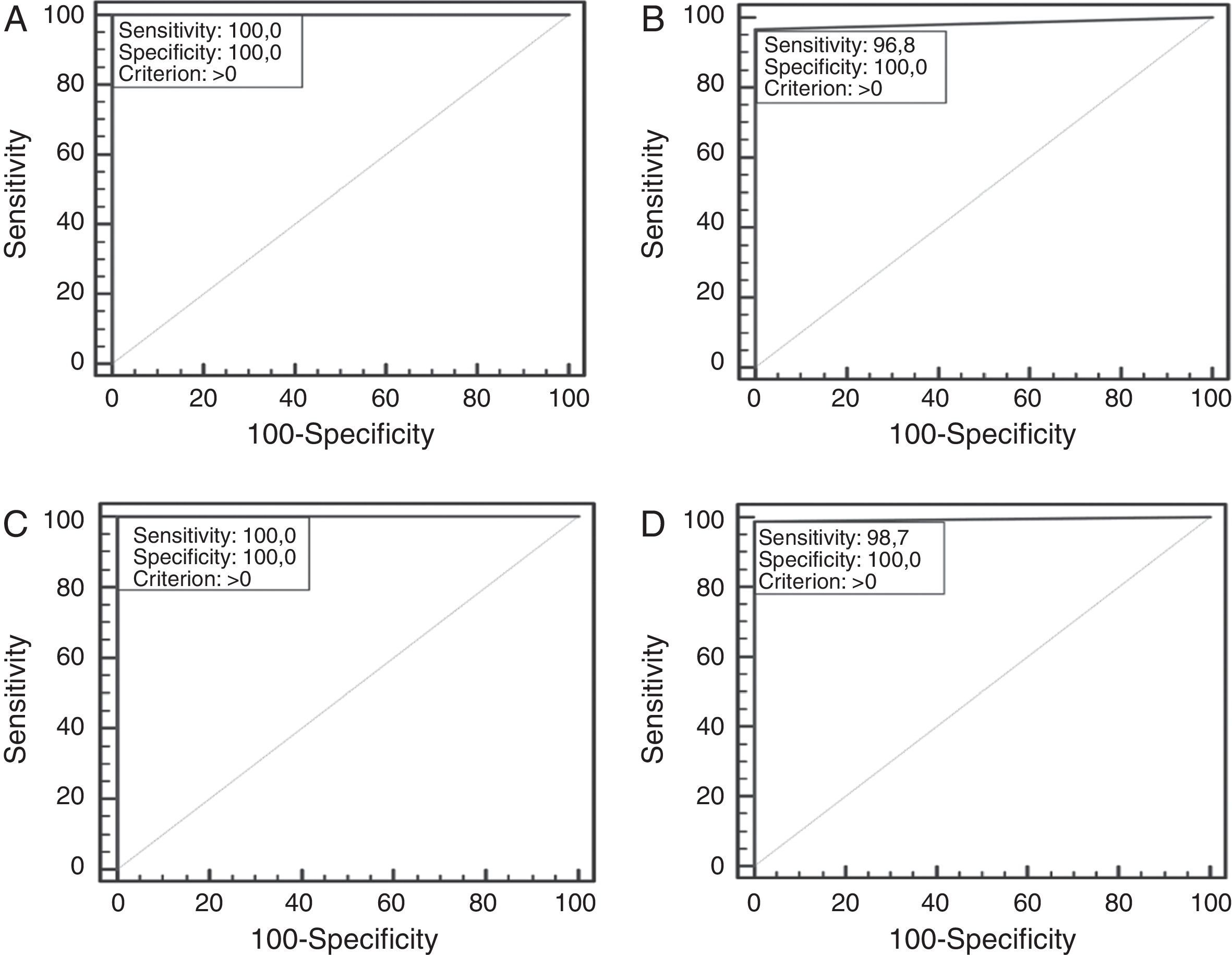
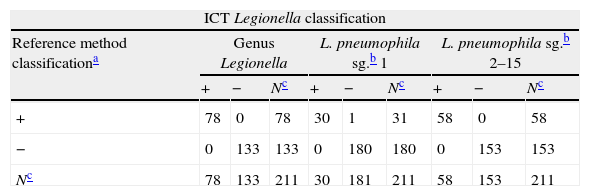
A total of 211 samples were used to analyze the performance of the ICT. They were bacteria strains from reference collections (NCTC, DSMZ, ATCC) (42L, 27LPN (13LPN1+14LPN2–15); 141 isolated from 64 water samples received by SGS Tecnos S.A. (Madrid, Spain) for Legionella testing (35L, 30 LPN (17 LPN1+13 LPN2–15), 106 non-Legionella (NOL)), which were characterized using a commercial latex slide test (Legionella Latex Test, Oxoid, UK) following the manufacturer‘s instructions; and 28 clinical isolates: 1 LPN1 Murcia9 (Vircell, S.L.’s collection) and 27 NOL from Hospital Neurotraumatológico de Jaén (Jaén, Spain) which were identified by biochemical panels (Microscan Walkaway, Siemens, Germany). All Legionella reference strains and the clinical isolated LPN1 Murcia were grown on a specific culture medium for Legionella, BCYE (Biomerieux, France), while blood-agar plates (Oxoid, UK) were used for the NOL clinical isolates. Environmental isolates samples were cultivated in parallel with both types of plate. One false negative result was obtained for the LPN1 test line (see Table 1) on the ICT. This specimen was re-analyzed by using direct immunofluorescence assays with a commercial mAb against LPN1 (clone Lp1 MAB 1 ATCC) and an in-house rabbit antiserogroup 1 anti-serum along with performing a commercial ELISA (Bartels, Ireland) for LPN1. The sample was negative in all three assays, suggesting that the commercial reference latex could have produced a false positive. Statistical analysis was performed with VassarStats (http://www.vassarstats.net). The Receiving Operating Characteristic (ROC) curves were generated with Medcalc® Software (http://www.medcalc.org/). As shown in Table 1, sensitivities of 100%, 100% and 96.7% were obtained for L, LPN and LPN1, respectively, and the overall sensitivity was 98.7%. The ICT showed a specificity of 100%. Predictive values, likelihood ratios, the Youden index and the accuracy provide optimal values for the classifier too. The ROC curves show the suitable performance of the classifier. The highest exactitude is achieved when the area under curve is 1, which was reached for the genus and the LPN2–15 lines; meanwhile, for the LPN1 line it was almost ideal (see Fig. 1).
Comparison of methods and statistical results for the ICT Legionella lines.
| ICT Legionella classification | |||||||||
| Reference method classificationa | Genus Legionella | L. pneumophila sg.b 1 | L. pneumophila sg.b 2–15 | ||||||
| + | − | Nc | + | − | Nc | + | − | Nc | |
| + | 78 | 0 | 78 | 30 | 1 | 31 | 58 | 0 | 58 |
| − | 0 | 133 | 133 | 0 | 180 | 180 | 0 | 153 | 153 |
| Nc | 78 | 133 | 211 | 30 | 181 | 211 | 58 | 153 | 211 |
| ICT Legionella statistical results* | ||||
| Genus Legionella | L. pneumophila sg.b 1 line | L. pneumophila sg.b 1–15 line | Overall | |
| Sensitivity | 100 (94.2–100) | 96.7 (81.4–99.8) | 100 (92.3–100) | 98.7 (92.1–99.9) |
| Specificity | 100 (96.5–100) | 100 (97.4–100) | 100 (96.9–100) | 100 (96.5–100) |
| PPVd | 100 (94.2–100) | 100 (85.9–100) | 100 (92.3–100) | 100 (94.1–100) |
| NPVe | 100 (96.5–100) | 99.4 (96.5–99.9) | 100 (96.9–100) | 99.2 (95.3–99.9) |
| ACf | 1 | 1 | 1 | 0.9953 |
| LRPg | Infinity | Infinity | Infinity | Infinity |
| LRNh | 0 | 0.0322 | 0 | 0.0128 |
| γi | 1 | 0.968 | 1 | 0.987 |
Summarizing, the evaluated ICT has shown suitable reliability and agreement with the reference methods. The ICT can identify Legionella genus and can differentiate between serogroup 1 and serogroups 2–15. The statistical results show a successful and non-random classification. So, the ICT can be considered a fast, easy-to-handle and reliable test for Legionella diagnosis in samples from cultures. It also represents a suitable confirmation method, due to its simplicity and high specificity and sensitivity values.
FundingThese studies have been fully funded by the company Vircell, S.L. (Santa Fe, Granada).
Conflict of interestTeresa Pardo, José Manuel Delgado and José Rojas are employees of Vircell, S.L. Antonio Fuertes works for SGS Tecnos, S.A. (Madrid) and has worked on the assignment and analysis of part of the samples evaluated.
All authors voluntarily publish this article and have no personal interest in these studies except that of disseminating scientific findings that have been planned and implemented, as well as monitoring, conducting research and analysis of results.