Heritability in schizophrenia can reach up to 80% and the risk in families is 5–10 times higher than in the general population. The large contribution of genetics in this disorder has led to a growing interest in its study.
ObjectivesTo review the findings of genetic studies known as Genomewide Association Studies (GWAS) on schizophrenia.
MethodSystematic search using Pubmed with the key words GWAS and (psychosis) or (schizophrenia). The following web pages have been reviewed: http://www.szgene.org/largescale.asp and www.genome.gov/gwastudies/.
ResultsThe GWAS have focused on causal biological aspects, such as the histocompatibility complex, glutamate metabolism, apoptosis and inflammatory processes, and the immune system (TNF-β, TNFR1). Also focused in the search were the genes that modulate the appearance of secondary metabolic and cardiac effects and secondary effects in subjects with schizophrenia and on anti-psychotic treatment. In neurorecognition, over-expression of the MET proto-oncogene (MET) has been associated with a low susceptibility for schizophrenia and a better cognitive performance, as well as a lower susceptibility for the incidence of cancer. Mention is also made of the different genes that mediate in cognitive functioning depending on the anti-psychotic treatment received.
ConclusionsThe main interests of the GWAS during the last few years have been the neurobiological pathways involved in schizophrenia. The discoveries arising from these studies have been limited. This has led to an innovative approach on the aetiological study of the disorder by studying gene–environment interactions.
La heredabilidad en la esquizofrenia llega hasta el 80% y el riesgo en familiares es 5–10 veces mayor que en la población general. La gran contribución genética en este trastorno ha llevado a dedicar un interés creciente a su estudio.
ObjetivosRevisar los hallazgos de los estudios genéticos conocidos como Genomewide Association Studies (GWAS) en esquizofrenia.
MétodoBúsqueda sistemática a través de Pubmed con las palabras clave GWAS and (psicosis) or (schizophrenia). Se han revisado las páginas http://www.szgene.org/largescale.asp y www.genome.gov/gwastudies/.
ResultadosLos GWAS se han centrado en aspectos biológicos causales como el complejo de histocompatibilidad, el metabolismo de glutamato, el proceso de apoptosis, inflamatorio y el sistema inmunológico (TNF-beta, TNFR1). También se han focalizado en la búsqueda de genes que modulen la aparición de efectos secundarios metabólicos, cardíacos y efectos secundarios en personas con esquizofrenia y tratamiento antipsicótico. En neurocognición se ha asociado la sobreexpresión del proto-oncogen MET (MET) con una baja susceptibilidad para la esquizofrenia y mejor rendimiento cognitivo, así como a menor susceptibilidad para la incidencia de cáncer. También se apuntado a diversos genes que medien en el funcionamiento cognitivo en función del tratamiento antipsicótico recibido.
ConclusionesLos principales intereses de los GWAS durante los últimos años han sido los recorridos neurobiológicos involucrados en la esquizofrenia. Los descubrimientos derivados de estos estudios han sido limitados. Esto ha conducido a un planteamiento innovador en el estudio etiológico del trastorno a través del estudio de las interacciones gen-ambiente.
Since 2005, multiple, ongoing, large-scale genetic studies have pinpointed most of the genes that make up the human genome. The objective of these genetic studies, known as Genome-Wide Association Studies (GWAS), is to link certain genes to different pathologies or disorders; as of the present time, certain common DNA sequences have been associated with susceptibility to more than 40 common diseases.1 Although new pathophysiological hypotheses have been formulated from these findings, it has not been possible to establish direct causal relationships between genetic markers and common pathologies.2
The first GWAS reports for psychiatric disorders appeared, and there are currently almost 100 GWAS in Psychiatry for ADHD, autism, bipolar disorder, schizophrenia, major depressive disorder, anxiety disorders, personality disorders, and neurocognition.
Despite the fact that GWAS interest in linking different genes to the risk of schizophrenia is growing and that about 1000 genes have been associated with a susceptibility to schizophrenia, it has not been possible to find a clear significance for any of them.3
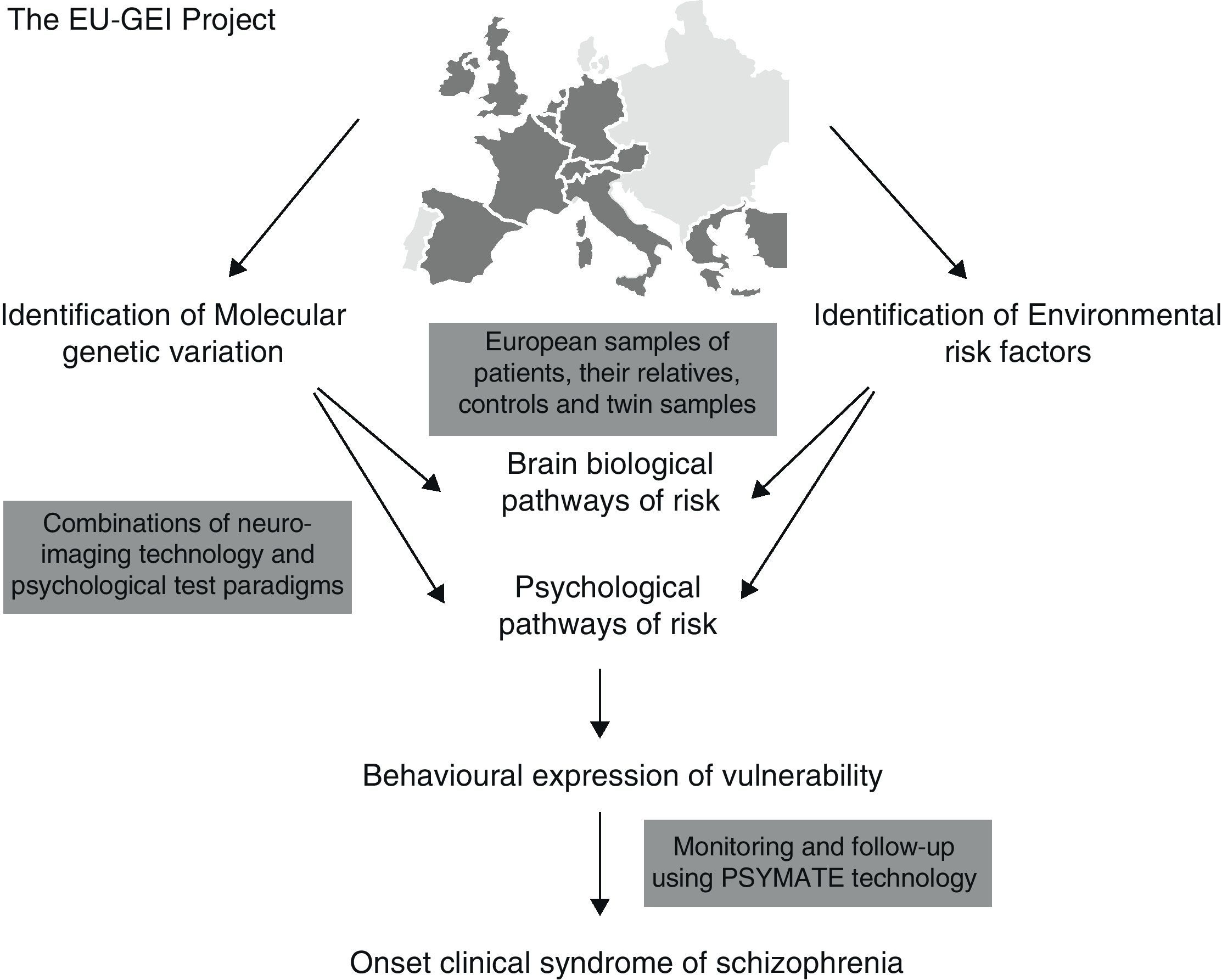
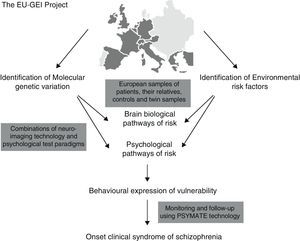
It has been proposed that epidemiological environmental studies be merged with genetic studies (GWAS) for the purpose of establishing new causal-environmental associations. A multi-centre project conducted by a network of multidisciplinary experts known as the European Network of Schizophrenia Networks for the Study of Gene Environment Interactions (EU-GEI)4 has been underway in Europe since 2010. The development, severity, and disease course of schizophrenia are being studied for the purpose of identifying the main causes and how these interact so that gene–environment interactions may be explored in depth and more fully understood. Spain is among the countries participating in this project, with locations in cities such as Barcelona, Madrid, Oviedo, Santiago de Compostela, and Valencia.
Fig. 1 shows implementation of the EU-GEI project.
The 2009 Psychiatric GWAS Consortium Coordinating Committee5 review of GWAS led to a proposal that all GWAS be systematically reviewed, along with their innovations in the field of schizophrenia, from the earliest studies to the present time.
Before describing the results found in the GWAS, we will give a simple definition of the genetic terms used throughout this review to make the content as clear as possible.
A gene is a linear nucleotide sequence on the DNA molecule containing the information required to synthesise a macromolecule having a specific cellular function; these are usually proteins, but they may also be mRNA, rRNA, and tRNA. The genome is the totality of genetic information an organism possesses. When speaking of the genome, we usually mean only the DNA content of the nucleus organised into chromosomes.
Chromosome is the name given to each of the little rod-shaped bodies in which the chromatin of the cell nucleus is organised during cell division (mitosis and meiosis). Chromatin is a microscopic substance made up of DNA that carries the genetic information for eukaryotic organisms.
In genetics, a deletion or suppression is a special type of chromosomal structural anomaly involving the loss of a DNA fragment from a chromosome, which may occur at the end of the chromosome (terminal) or along one of its arms (interstitial). A duplication is the repetition of a chromosome fragment below the original fragment. A single-nucleotide polymorphism or SNP (pronounced “snip”) is a DNA sequence variation affecting only 1 base of a genome sequence (adenine [A], thymine [T], cytosine [C], or guanine [G]). However, some authors believe that changes of a few nucleotides, such as little insertions and deletions (indels), may be considered SNPs, where the term simple nucleotide polymorphism is more appropriate. Such a variation must occur in at least 1% of the population to be considered an SNP; at less than 1%, it is considered a specific mutation rather than an SNP. Copy-number variations (CNV) are alterations of the DNA of a genome that result from having an abnormal number of copies of one or more sections of the DNA. CNVs correspond to relatively large regions of the genome that have been suppressed or amplified on certain chromosomes.
MethodThis review systematically describes the articles on schizophrenia published in GWAS. PubMed was searched on the keywords GWAS and (psychosis) or (schizophrenia); web page http://www.szgene.org/largescale.asp was also consulted, which lists all the GWAS publications on schizophrenia as well as other non-GWAS articles from large-scale studies on schizophrenia, along with www.genome.gov/gwastudies/ to compare the results and studies linked to the GWAS that have been cited in this review.
Using the keywords GWAS and schizophrenia, 55 articles were found, of which 12 were reviews and 43 were original articles published between 2009 and 2011; the keywords GWAS and psychosis yielded 8 articles, 7 of which had already appeared in the previous search.
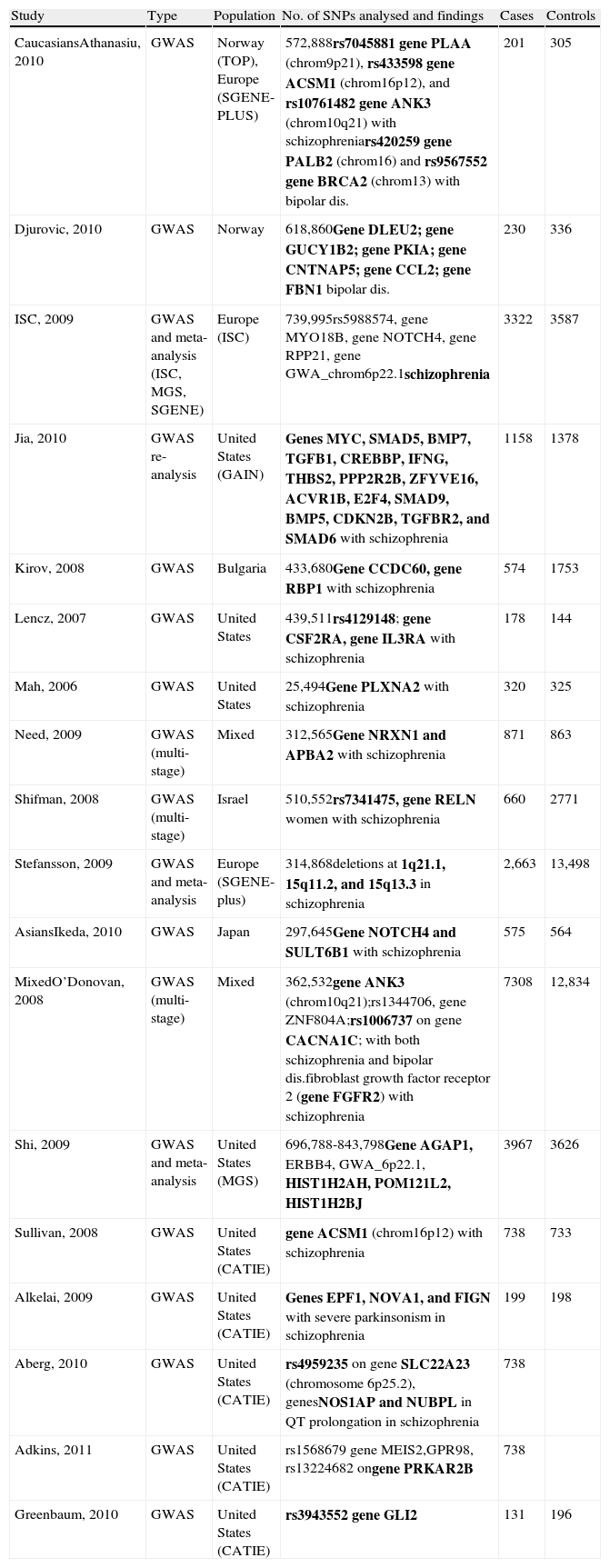
Table 1 summarises all the GWAS on schizophrenia that have been published to date.
Summary of GWAS on schizophrenia.
| Study | Type | Population | No. of SNPs analysed and findings | Cases | Controls |
| CaucasiansAthanasiu, 2010 | GWAS | Norway (TOP), Europe (SGENE-PLUS) | 572,888rs7045881 gene PLAA (chrom9p21), rs433598 gene ACSM1 (chrom16p12), and rs10761482 gene ANK3 (chrom10q21) with schizophreniars420259 gene PALB2 (chrom16) and rs9567552 gene BRCA2 (chrom13) with bipolar dis. | 201 | 305 |
| Djurovic, 2010 | GWAS | Norway | 618,860Gene DLEU2; gene GUCY1B2; gene PKIA; gene CNTNAP5; gene CCL2; gene FBN1 bipolar dis. | 230 | 336 |
| ISC, 2009 | GWAS and meta-analysis (ISC, MGS, SGENE) | Europe (ISC) | 739,995rs5988574, gene MYO18B, gene NOTCH4, gene RPP21, gene GWA_chrom6p22.1schizophrenia | 3322 | 3587 |
| Jia, 2010 | GWAS re-analysis | United States (GAIN) | Genes MYC, SMAD5, BMP7, TGFB1, CREBBP, IFNG, THBS2, PPP2R2B, ZFYVE16, ACVR1B, E2F4, SMAD9, BMP5, CDKN2B, TGFBR2, and SMAD6 with schizophrenia | 1158 | 1378 |
| Kirov, 2008 | GWAS | Bulgaria | 433,680Gene CCDC60, gene RBP1 with schizophrenia | 574 | 1753 |
| Lencz, 2007 | GWAS | United States | 439,511rs4129148; gene CSF2RA, gene IL3RA with schizophrenia | 178 | 144 |
| Mah, 2006 | GWAS | United States | 25,494Gene PLXNA2 with schizophrenia | 320 | 325 |
| Need, 2009 | GWAS (multi-stage) | Mixed | 312,565Gene NRXN1 and APBA2 with schizophrenia | 871 | 863 |
| Shifman, 2008 | GWAS (multi-stage) | Israel | 510,552rs7341475, gene RELN women with schizophrenia | 660 | 2771 |
| Stefansson, 2009 | GWAS and meta-analysis | Europe (SGENE-plus) | 314,868deletions at 1q21.1, 15q11.2, and 15q13.3 in schizophrenia | 2,663 | 13,498 |
| AsiansIkeda, 2010 | GWAS | Japan | 297,645Gene NOTCH4 and SULT6B1 with schizophrenia | 575 | 564 |
| MixedO’Donovan, 2008 | GWAS (multi-stage) | Mixed | 362,532gene ANK3 (chrom10q21);rs1344706, gene ZNF804A;rs1006737 on gene CACNA1C; with both schizophrenia and bipolar dis.fibroblast growth factor receptor 2 (gene FGFR2) with schizophrenia | 7308 | 12,834 |
| Shi, 2009 | GWAS and meta-analysis | United States (MGS) | 696,788-843,798Gene AGAP1, ERBB4, GWA_6p22.1, HIST1H2AH, POM121L2, HIST1H2BJ | 3967 | 3626 |
| Sullivan, 2008 | GWAS | United States (CATIE) | gene ACSM1 (chrom16p12) with schizophrenia | 738 | 733 |
| Alkelai, 2009 | GWAS | United States (CATIE) | Genes EPF1, NOVA1, and FIGN with severe parkinsonism in schizophrenia | 199 | 198 |
| Aberg, 2010 | GWAS | United States (CATIE) | rs4959235 on gene SLC22A23 (chromosome 6p25.2), genesNOS1AP and NUBPL in QT prolongation in schizophrenia | 738 | |
| Adkins, 2011 | GWAS | United States (CATIE) | rs1568679 gene MEIS2,GPR98, rs13224682 ongene PRKAR2B | 738 | |
| Greenbaum, 2010 | GWAS | United States (CATIE) | rs3943552 gene GLI2 | 131 | 196 |
In schizophrenia, heritability is as high as 80%, and the relatives of those with schizophrenia are at a 5–10 times greater risk of suffering from it than the general population.6 Individuals with schizophrenia have a significantly higher probability of deletions and duplications7 in their genome.
The most recent GWAS have postulated that schizophrenia, autism, and bipolar disorder have genetic origins in common.8 Through genome sequencing technology, studies of structural genetic variation, such as copy-number variations (CNV), have been joined with rare genetic variations to complement gene–gene interaction studies. Most CNVs are single ones on chromosomes 1q21.1, 15q11.2, 15q13.3, 16p11.2, 17p12, or 22q11.2,7,9–11 which have also been implicated in autism and other developmental disorders.10 Duplications and perhaps also suppressions on chromosome 16p13.1, previously associated with autism and mental retardation, have also been associated with a risk of schizophrenia.12
A GWAS study conducted in Norway13 with a sample of 210 cases and 305 controls associated schizophrenia with simple nucleotide polymorphisms (SNP) rs7045881 on chromosome 9p21; rs433598 on chromosome 16p12; and rs10761482 on chromosome 10q21. These markers were located on genes PLAA, ACSM1, and ANK3, respectively. Gene PLAA (on chromosome 9) had not previously been described as a susceptibility gene, but chromosome 9p21 had already been implicated as a linkage region for schizophrenia. Gene ACSM1 (on chromosome 16) had previously been identified in the CATIE study14 as a susceptibility gene for schizophrenia, and gene ANK3 (on chromosome 10) has been associated with both schizophrenia and bipolar disorder, which supports the hypothesis of overlapping genetic susceptibility between these psychopathologies.8,15 The SNP rs1344706 on gene ZNF804A (which codes for protein 804A) and the SNP rs1006737 on gene CACNA1C on chromosome 12 (which codes for subunit alpha 1C) have also been associated as regions related to both schizophrenia and bipolar disorder,15 with a possible gender modulation of the risk for schizophrenia having been indicated recently.16 These last results were replicated by the Cardiff group itself in 201017,18; more recently, however, the Schanze et al. group were unable to replicate the association between gene ZNF804A and schizophrenia.19
The same Norwegian sample was used to investigate the connection between genetic variations in SNPs rs420259 on gene PALB2 (chromosome 16) and rs9567552 on gene BRCA2 (on chromosome 13)—variations associated with intracellular functions of expressed proteins in breast cancer—and schizophrenia and bipolar disorder. Analysis of a sample of 781 patients with schizophrenia and 686 with bipolar disorder revealed that these genetic variations were associated with bipolar disorder but not with schizophrenia.20
In another study conducted in the United States in 2010,21 GWAS data was combined with data from 3 large efficacy studies, including a sample of individuals with schizophrenia (CATIE, n=741), with bipolar disorder (STEP-BD, n=1575), and with major depressive disorder (STAR*D, n=1204). The SNP rs6484218, near the adrenomedullin gene on chromosome 11p15, was associated with bipolar II disorder but not with the rest of the psychiatric disorders. In a recently published European GWAS study,22 variations in different genes on chromosome 11 (AMBRA1, DGKZ, CHR, and MDK) were also associated with schizophrenia. Its highly standardised and homogeneous sample included 1169 patients (464 from Germany, 705 from Holland) and 3714 ethnicity-matched controls. A follow-up study was subsequently conducted on 2569 individuals with schizophrenia and 4088 controls (from Germany, Holland, and Denmark). These findings were again replicated in 23,206 independent samples from European ancestors (P=.0029, OR=1.11). In a neuroimaging study done on the sample, healthy carriers of the risk allele showed altered activation in the cingulate cortex during cognitive control tasks. The area of interest corresponded to an interface between emotional regulation and cognition which, in individuals with schizophrenia and bipolar disorder, is both functionally and structurally abnormal.
In some genetic studies, SNPs rs7683874 and rs10937823 on gene SORCS2 of chromosome 4p15-p16 have also been identified as candidate regions for the risk of bipolar disorder and schizophrenia.23
Glutamate metabolism, the process of apoptosis, the inflammatory process, and the immune system (TNF-beta, TNFR1) are signalling pathways that have been associated with schizophrenia.24 The O’Donovan et al. GWAS study,25 with a sample of 5142 cases of schizophrenia and 6561 controls from the United States, Australia, Germany, China, Japan, Israel, and Sweden, found evidence of association between schizophrenia and the SNP rs17101921 near the genetic region that codes for fibroblast growth factor receptor 2 (FGFR2). This growth factor is associated with an increased mitotic activity index and DNA synthesis, facilitating the proliferation of various precursor cells, such as chondroblasts, collagenoblasts, and osteoblasts, for example, that form the body's fibrous, connective, and support tissues. This factor has also been associated with tumour angiogenesis in oncogenic processes.
Both schizophrenia and bipolar disorder as well as other psychiatric disorders, such as autism, specific language disorders, and learning difficulties, have been connected with alterations in a series of genes (NRXN1, CNTNAP2, and CASK) associated with cell adhesion molecules (CAM), which would be responsible for synapse formation and normal cell transmission.26,27
GWAS have been conducted in different ethnic populations to look for differences in genetic susceptibility to schizophrenia. A recent, family-based GWAS project28 that included a sample of 107 Israeli Jewish families, found an association between gene DOCK4 (SNP rs2074127, P=1.134×10−7) and 6 additional significant associations (P<1×10−5). One of the SNPs (rs4803480) located on gene CEACAM21 was replicated significantly in a family-based sample of Arab-Israeli origin (P=.002). Significant associations were also found with genes PGBD1, RELN, and PRODH, replicating the results previously obtained in other studies.
In a GWAS study conducted on an Asian population,29 with a Japanese and Chinese cohort of 2535 individuals, the most significant value was for an SNP in gene ELAVL2 (embryonic lethal, abnormal vision, Drosophila-like 2) on chromosome 9p21.3 (P=.00087).
Findings on the inducement of motor, cardiac, and metabolic side effectsThere is a high incidence of side effects in individuals who are on antipsychotic therapy. Antipsychotics-induced parkinsonism is a severe adverse effect of neuroleptic therapy. Factors such as the type of treatment, being elderly, and being female have been associated with a greater risk of having side effects, but it is obvious that individuals vary in their susceptibility to them. This is why the GWAS have been focused on studying some genes that may influence the appearance of side effects.
The CATIE study included a GWAS analysis focused on this type of side effects,30 and a sample of 199 Americans with schizophrenia was analysed. The sample was randomised to antipsychotic monotherapy and followed for a period ranging from 2 weeks to 18 months during the first phase of the CATIE study. Parkinsonian effects were monitored, and genes EPF1, NOVA1, and FIGN were identified as candidate genes associated with severe parkinsonism secondary to this therapy.
A sample of 738 individuals with schizophrenia, also from the CATIE study, was analysed to explore the association of some antipsychotics with prolongation of the QT interval and the risk of cardiac arrhythmias. In the case of quetiapine, the presence of SNP rs4959235 on gene SLC22A23 (chromosome 6p25.2) was associated QT prolongation.31
An Israeli subsample from the CATIE study32 was used to analyse candidate genes that may have influenced the appearance of tardive dyskinesia in individuals with chronic schizophrenia; it included 327 individuals with schizophrenia who were on antipsychotic therapy. They analysed about 495,000 simple nucleotide polymorphisms and associated the SNP rs3943552 on gene GLI2 with tardive dyskinesia—a fact that had already been observed in subsamples of Jewish patients from Ashkenazi.
Lastly, the genetic variation that may affect susceptibility to metabolic side effects has been studied. In the same sample from the CATIE study,33 21 polymorphisms were associated with metabolic side effects, such as weight gain induced by antipsychotic medication, blood lipid profile, blood glucose and haemoglobin A1C, blood pressure, and cardiac ratio. Gene MEIS2 on chromosome 15q14 mediated the effects of risperidone on not only hip circumference (q=0.004) but also waist circumference (q=0.055), besides having secondary associations with the BMI and systolic and diastolic pressure. Another significant finding was with gene GPR98 (on chromosome 5q14.3), which acted as mediator in the effects of risperidone on haemoglobin A1C levels. Other genes mediating metabolic effects were PRKAR2B, FHOD3, RNF144A, ASTN2, SOX5, and ATF7IP2.
Body mass index (BMI) has been widely studied in GWAS, and although more than a dozen variations have been associated, individually they account for only a small proportion of the variance. For this reason, calculation of the genetic risk sum score (GRSS) has been proposed, which encompasses the total risk alleles discovered. Peterson et al.17 took a sample of 2653 Caucasians and 973 African-Americans corresponding to 2 meta-analyses and calculated the GRSS with covariables that affect BMI (age, sex, ancestry, and the like). A strong association with the BMI was demonstrated (P=3.19×10−6), but it accounted for only a limited portion of the variance (0.66%). The GRSS and covariables were strong predictors for the overweight and obesity categories, but the maximum discrimination was for type III obesity (AUC=0.697).
Findings in neurocognitionIn the field of neurocognition in schizophrenia, GWAS have focused their efforts on identifying genetic variations involved in cognitive modulation. These studies have proposed the use of functional neuroimaging techniques, such as quantitative phenotyping, in combination with GWAS, which represents a step toward improved understanding of the cognitive aspects of schizophrenia. These studies are based on measuring consumption of blood oxygen (BOLD) in the dorsolateral prefrontal cortex during a working memory task evaluation (using the Sternberg paradigm) as well as in combination with genome scanning. This technique may provide greater statistical power than each test has separately.34
The 2007 Hippisley-Cox et al.35 GWAS studied the risk of 6 common cancers in individuals with schizophrenia. In a sample of 40,441 individuals with cancer, 107 Caucasians with schizophrenia were found and compared with 112 healthy controls, genotyping the effect of the MET variation. When cognitive functioning was explored in a sample of 191 individuals with schizophrenia and 188 healthy controls, a significant association was reported between the MET proto-oncogene and susceptibility to having schizophrenia. The presence of 2 or more copies of haplotype GCAATACA was associated with reduced probability of developing schizophrenia, compared to subjects who did not have copies. A significantly positive impact on neurocognition was reported for those who were carriers of genotype METGCAATACA.
Various complexin 2 gene polymorphisms have been associated with modification of cognitive performance in individuals with schizophrenia, compared with the control population. A sample of 1037 patients with schizophrenia and 2265 healthy controls was analysed in a multi-centre study conducted at 23 psychiatric centres in Germany.36 The primary parameters of cognitive performance were evaluated, including measurement of executive functioning, reasoning, verbal learning, and memory. Six polymorphisms distributed across the complexin 2 gene proved to be strongly associated with the schizophrenic subjects’ current cognition but only marginally associated with their premorbid intelligence. These studies, where subjects were examined by a single research team, gave rise to the Göttingen Research Association for Schizophrenia (GRAS) database.37 Created in Germany from data collected by a single research team, the GRAS aims to be the foundation for studying genetic causes of the schizophrenic phenotype in a “phenotype-based genetic association (PGA) study.” It is important to highlight that, while these are not GWAS studies, their results complement those obtained through genome-wide association studies (GWAS) on schizophrenia.37
In a study using a sample of 738 patients with schizophrenia taken from the CATIE study38 and grouped according to which of 5 antipsychotic therapies they were receiving (olanzapine, perphenazine, quetiapine, risperidone, and ziprasidone), patients completed different neurocognitive batteries measuring processing speed, verbal memory, vigilance, reasoning, and various working memory domains. Six SNPs, located in the proximity of genes EHF, SLC26A9, DRD2, GPR137B, CHST8, and IL1A, were identified as cognitive mediators. Neurocognitive scores were also more strongly associated with SNP rs286913 on gene EHF (P=6.99×10−8), mediating the effects of ziprasidone on vigilance; SNP rs11240594 on gene SLC26A9 (P=1.4×10−7), mediating the effects of olanzapine on processing speed; and SNP rs11677416 on gene IL1A (P=6.67×10−7), mediating the effects of olanzapine on working memory.
Lastly, we highlight that incorporating neuroimaging and optical manipulation studies into GWAS studies has made it possible to implicate the genes coding for FGF17 and glypican-1 in the brain development of the negative symptoms of schizophrenia.39
Practical aspects of conducting GWASThere are articles that analyse the numerous GWAS studies’ methodology.5,40–42 These articles place special emphasis on the importance of family studies because they demonstrate shared familial responsibility through the diagnostic limits (in both schizophrenia and bipolar disorder).40
Sanders et al.43 propose the use of technology via the Internet to create a database of controls that could be shared among researchers, enabling them to select proper controls for this type of study.43
A meta-analysis has recently been conducted44 in accordance with 2 different methods—genome scan meta-analysis (GSMA) and Badner and Gershon's multiple scan probability method (MSP)—applied to 13 and 16 databases for bipolar disorder and schizophrenia, respectively. The authors found that the 2 methods yield different genomic regions, which suggests that, in this modern era of genomics, comparative linkage meta-analyses (CLMA) could be used to optimise the discovery of rare and low-frequency variations. Another option for improving the findings from GWAS studies is the SNP-based pathway enrichment method.45 There are 2 steps to this method: first, using a truncated statistical product to identify all representative SNPs for each gene, to calculate the mean representative SNPs for the genes, and to select those that are representative based on their position. Next, they are put in order by statistical significance of their association with the trait of interest, and a Kolmogorov–Smirnov test is applied.
With regard to the techniques used in GWAS, the haplotype test has been proposed, which uses a model to analyse multimarkers for rare variations in candidate regions that could be related to the appearance of schizophrenia.46
DiscussionDiscoveries from GWAS studies and genetic studies related to the GWAS focus on different aspects of schizophrenia. In the purely genetic realm, genetic variations on different common chromosomes have been described for different pathologies, such as schizophrenia, bipolar disorder, and autism. All these findings point to a possible common aetiology for different psychiatric disorders, such as schizophrenia and bipolar disorder.2
In schizophrenia, many findings have been related to (1) different signalling pathways for development of proteins (fibroblasts, chondroblasts) participating in body support functions and in tumour angiogenesis in oncogenic processes, as well as (2) signalling pathways for cell adhesion molecules (CAM) responsible for synapse formation and normal cell transmission. This has enabled new hypotheses on the cause of schizophrenia to be set forth, with models proposing multi-system involvement and not solely psychiatric involvement, as an author had previously suggested.47,48
Understanding how genes modulate the appearance of adverse side effects may help to improve classification and knowledge of them as well as prevent their appearance. Improved knowledge of neurocognition in schizophrenia and how it is linked to genetic alterations may enable us to better understand our patients’ cognitive functioning and make cognitive rehabilitation therapies more purposeful.
The limitations of GWAS studies are basically related to the large-scale genomic research. These studies do not allow for searching on a single gene or chromosome associated with greater susceptibility, which is why the International Schizophrenia Consortium (ISC) has recently sought other alternatives. They have reported that using analysis methods like the polygenic score method could detect marginal associations between susceptibility alleles for schizophrenia and other SNP (eQTL) affecting genetic expression, and that individuals who have this marginal association with SNP cis-eQTL could be at higher risk for schizophrenia than those who do not have it.49 Another limitation of GWAS studies is related to the high cost they entail. Their sometimes questionable cost-benefit has led to new approaches in which the genetic results of GWAS studies are merged with those of environmental or epidemiological studies.
ConclusionsAlthough GWAS studies of recent years have opened up new lines of research, it has not been possible to conclude from them specific knowledge that may be directly applied in daily clinical practice. These limitations have led some researchers to contemplate a multidisciplinary project merging genetic studies with environmental studies. This is how the European Network of Schizophrenia Networks for the Study of Gene–Environment Interactions (EU-GEI)7 project began, in which various disciplines complement and are indispensable to each other in studying gene–environment interactions. This proposal may yield promising results.
In the coming years, new studies must be developed that would include meta-analyses of more than 10,000 subjects2; delve into the nature of environmental factors; and enable the gene–environment interaction, the nature of genetic variation, and the mechanism of gene–environment interactions to be explained.50
This new approach may represent a step forward in our knowledge of schizophrenia, enabling us to better define the disorder, understand its aetiology, and discover new treatment targets for reducing sequelae and improving symptomatic management in our patients.
Conflicts of interestThe authors have no conflicts of interest to declare.
Please cite this article as: Valiente A, et al. Revisión sistemática de los Genomewide Association Studies (GWAS) en esquizofrenia. Rev Psiquiatr Salud Ment (Barc.). 2011;4:218–27.