In this study, we describe two closely related cases of multifocal MSSA infections within the same household. The dissemination of infection was severe in both cases, but none of the patients presented endocarditis. All isolates shared resistance to erythromycin and clindamycin (inducible macrolide resistance phenotype, iMLSb). The genomic analysis of one of the isolates indicated that it belongs to the human-associated CC398-MSSA clonal complex, an emerging lineage in our region. Whole genome sequencing enabled the characterization and differentiation of this clone from the livestock-associated CC398-MSSA lineage.
En este estudio, describimos dos casos estrechamente relacionados de infecciones multifocales por SAMS en personas convivientes. La diseminación de la infección fue severa en ambos casos, pero ninguno de los pacientes presentó endocarditis. Todos los aislamientos compartieron el fenotipo de resistencia a eritromicina y clindamicina (fenotipo inducible iMLSb). El análisis genómico de uno de los aislamientos indicó que pertenece al CC398-SAMS asociado a humanos, un linaje emergente en nuestra región. La secuenciación del genoma completo permitió caracterizarlo y distinguirlo del linaje CC398-SAMS asociado al ganado.
Staphylococcus aureus (SA) is one of the most commonly isolated pathogens responsible for a wide range of infections including moderate skin conditions, but also life-threatening diseases such as pneumonia, bacteremia and disseminated infections, depending on the susceptibility of the host and SA pathogenic factors.
Outbreaks of severe community-associated skin and soft tissue infections are often linked to epidemic and pandemic methicillin-resistant S. aureus (CA-MRSA), whereas outbreaks involving methicillin-susceptible S. aureus (MSSA) are rarely described. Here, we report two closely related cases of severe multifocal MSSA infections within the same household, associated with CC398-MSSA.
Patient 1In February 2023, a 70-year-old woman arrived at the emergency department with a seven-day history of erythema and pain in the right thigh, following an intramuscular injection. Twelve hours later, she developed an erythematous nodule in the right sternoclavicular region, accompanied by fever and night sweats. A month before, she had undergone mastectomy for breast cancer.
Initial evaluation revealed neutrophilia (leukocytes 22000/ml, neutrophils 86%) and an elevated creatinine level (1.88μmol/l). A large right gluteal abscess was drained, and antibiotic treatment with vancomycin was initiated. Sternoclavicular joint toileting was also performed. Cefazolin was added to the treatment after detecting Gram-positive cocci in clusters in the drained material and initial blood cultures. Upon confirmation of monomicrobial MSSA bacteremia, vancomycin was interrupted, and treatment continued with cefazolin 6g/day.
MSSA isolates recovered from all samples shared the same susceptibility profile, exhibiting resistance to erythromycin and clindamycin (inducible macrolide resistance phenotype, iMLSb). Control blood cultures 48h after the start of treatment revealed S. aureus, and this result was repeated in one of two bottles after seven days. Echocardiogram showed no evidence of endocarditis. On day 9, due to persistent fever, a tomography revealed contralateral gluteal abscess, spondylodiscitis with epidural involvement, and a psoas abscess. Drainage of the gluteal abscess was performed, while a conservative approach was taken for the psoas and epidural abscesses. Control blood cultures after 9 days of treatment were negative. Ten days later, spinal MRI showed worsening of the epidural and psoas abscess, prompting drainage of the epidural collection. Another sternoclavicular joint toileting was performed due to persistent swelling. There was no evidence of bone involvement. The patient was discharged after completing 28 days of intravenous antibiotic treatment, following negative blood cultures.
Patient 2In early June 2023, patient 1's partner, a 78-year-old man, presented to the hospital with fever and swelling in the right trochanteric region of the right hip. He experienced pain in the left thigh, starting 48h after acupuncture for interscapular pain. Diagnosed with acute myeloid leukemia during his wife's hospitalization in February, he had been receiving venetoclax and azacitidine until 21 days before admission. In addition, he had a history of total right knee replacement in 2018.
On examination, the patient was drowsy, with a temperature of 38.5°C, pulse rate of 110/min, chills, normal blood pressure, and partial disorientation. No murmurs or signs of endocarditis were observed. A collection in the right trochanteric region was drained, yielding abundant purulent drained fluid. Laboratory findings revealed 6610WBC/mm3, low platelet count (67000/mm3), and elevated inflammatory markers (erythrocyte sedimentation rate >120mm/h and C-reactive protein 288mg/l). Treatment with vancomycin 1g/8hs and piperacillin–tazobactam was initiated. Soft tissue ultrasound confirmed another collection (73mm×33mm) in the left inner thigh, which was drained the same day.
The next day, a fluctuating area in the left scapular region, nodules in the left leg, swelling in the left hallux and swelling and pain in the right prosthetic knee were observed. MSSA, resistant to erythromycin and clindamycin (showing iMLSb phenotype), was obtained from blood cultures and purulent drained fluid. Treatment was modified to cefazolin, and an echocardiogram showed no evidence of endocarditis. One day after admission, arthritis was confirmed by arthrocentesis, and the patient underwent arthroscopy and drainage of multiple abscesses in the right iliac region, right thigh, two in the left thigh, and another in the left extrapleural thoracic region. Control blood cultures 48h after beginning treatment were negative.
Five days later, rifampicin was added for the prosthetic infection with prosthesis conservation. On day 6, a transesophageal echocardiogram was performed without evidence of endocarditis. Due to persistent fever on day 8, a tomography of the abdomen and chest was performed, ruling out other secondary foci. After 14 days of fever without evident foci, daptomycin was added. The patient became afebrile and was transferred to complete treatment in another hospital.
Given the identical phenotype and clinical similarities in both severe disseminated multifocal MSSA infections without infective endocarditis and considering that both patients came from the same household, we recognized the importance of studying the strain obtained from the second patient, as the isolate recovered from the first patient had already been discarded.
The isolate obtained from patient 2, HSCB1, was identified by conventional methods and confirmed by MALDI-TOF MS (Bruker Daltonics) as S. aureus. DNA was extracted using the Wizard® Genomic DNA purification kit (Promega, Madison, MI, USA). Shotgun gDNA libraries were prepared and whole genome sequencing was performed on the Illumina NovaSeq 6000 platform with 150bp paired-end reads. Quality control was performed using FastQC (version 0.12.1/Galaxy Version 0.74+galaxy0, https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and Kraken2 (Galaxy Version 2.1.3+galaxy1, https://doi.org/10.1186/s13059-019-1891-0). Multilocus sequence type (MLST) was determined with MLST (Galaxy Version 2.22.0, https://github.com/tseemann/mlst) and the spa type with SpaTyper 1.0 from the Center for Genomic Epidemiology. Antibiotic resistance and virulence determinants were detected using ABRicate (Galaxy Version 1.0.1, https://github.com/tseemann/abricate) with Resfinder (2020-Apr-19)-NCBI (2020-Apr-19, https://www.ncbi.nlm.nih.gov/pathogens/refgene/) databases and VFDB (2020-Apr-19, https://academic.oup.com/nar/article/47/D1/D687/5160975)-VirulenceFinder (https://cge.food.dtu.dk/services/VirulenceFinder/citations.php) databases, respectively. Plasmid replicons were determined using Plasmidfinder (2020-Apr-19, https://github.com/genomicepidemiology/plasmidfinder) using ABRicate. Whole genome phylogeny was inferred after mapping reads to the S0385 (accession AM990992.1) reference genome using the snp phylogeny tool as previously described5, to determine the genetic relatedness between HSCB1 and 55 public CC398 S. aureus genomes (Supplementary table). Forty-five genomes correspond to clinical isolates from South America, and the remaining 10 from different countries in North America and Europe. Most of these 10 genomes belong to clinical samples, while two were recovered from environmental isolates from Hawaii (Supplementary table).
HSCB1, was mecA negative, belonged to CC398, and spa type t1451 (CC398-MSSA-t1451). Regarding antimicrobial resistance determinants, it harboured the ermT gene, which is associated with the iMLSb phenotype for erythromycin–clindamycin and with replicon rep13_2. HSCB1 was negative for Panton Valentine Leukocidin genes (lukS/F-PV), but it harboured the immune evasion cluster (scn, chp) on the ϕSa3 prophage, a feature of the CC398 human lineage10. Additionally, we found other virulence determinants such as genes related to surface adhesion (ebp, clfA, spa, map, clfB, fnbA, fnbB, sbi, sdrC, sdrE), hemolysis (hly/hla, hlb, hld, hlgA-C), iron acquisition (isd operon), biofilm formation (icaA-D), capsule formation (cap8A-G), type VII secretion system (esaA-B, essA-B), and diverse enzymes (coa, geh, lip, hysA, sspA, sspB, sspC, aur, adsA).
Historically, MSSA clones have been poorly studied despite causing the majority of S. aureus infections globally. Little is known about their clonal evolution, but advances in whole genome sequencing technologies have shed more light on the issue5.
The evolutionary history and the international spread of CC398 has been previously described10. First, a livestock-associated CC398-MRSA clone (LA-CC398) was identified causing infections in farmers. Later, a CC398-MSSA clone capable of exclusively infecting humans or livestock independent (Hu-CC398) was described and initially spread to some European countries, the Caribbean and North America1,3,8. In China it was highly prevalent, causing 20% of skin and soft tissue infections2. Further studies linked CC398 to the increasing iMLSb resistance rates found in MSSA during the last years in our region5,7.
Evidence suggests that the Hu-CC398 lineage is exclusively transmitted among humans2,4 using an optimized repertoire of adhesion molecules9. Cases of colonization in cohabiting households, such as the one in this report, have been also previously described9. Environmental surfaces can be effective reservoirs for CC398 transmission6. Hu-CC398, harboring the ermT gene and the prophage ϕSa3 carrying genes involved in human-specific immune evasion cluster2,8,9, causes many invasive infections such as bacteremia, endocarditis and bone joint infections2.
The complexity and invasiveness of the clinical cases described herein can be associated with the patients’ previous health conditions, the remarkable number of virulence factors of this pathogen and the particular features of this clonal complex. In addition, the observation of apparent transmission between patients living together in the same household reflects a significant concern, supported by evidence found in the literature2.
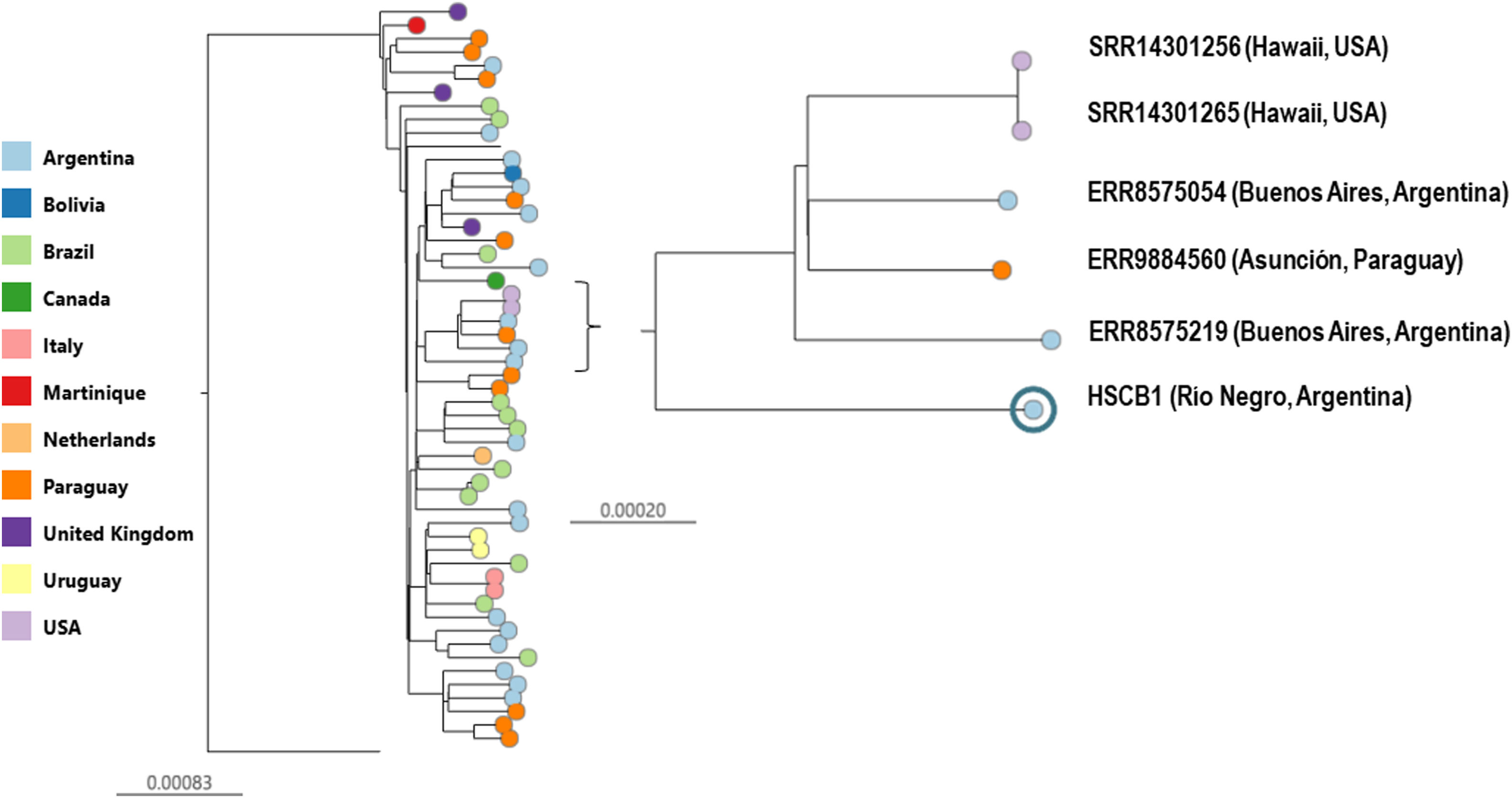
The phylogenetic analysis showed that HSCB1 clustered with other Hu-CC398 genomes and was closely related to three invasive clinical strains recovered in our region from Buenos Aires city, and Paraguay5 in 2019 and two environmental isolates recovered from Hawaii in 20206 (98.4/100% bootstrap support, Fig. 1).
Whole genome phylogeny of methicillin-susceptible Staphylococcus aureus CC398 genomes. S0385 (reference)-rooted maximum likelihood phylogenetic tree inferred from 4176 SNPs showing the genetic relationships among CC398-MSSA isolated from South America, North America and Europe. Tree nodes are coloured by country of isolation as indicated in the legend. Scale bars represent the number of single nucleotide polymorphisms (SNPs) per variable site. A specific clade of interest containing the genome in study (98.4/100% bootstrap support) is zoomed-in.
Interestingly, these two isolates were recovered from water samples in Hawaii, showing the ability of these strains to persist in the environment and successfully spread across different geographical and environmental contexts.
Our study documented highly severe clinical cases of MSSA infections associated with CC398 in Argentina, and suggested a possible transmission of CC398 within cohabiting individuals (same antibiotic susceptibility profile in both isolates), highlighting the importance of prevention and control measures in contexts where close contact of environmental exposure facilitates its transmission. These results are a contribution to the understanding of the high epidemicity and pathogenicity of this clonal complex.
The raw sequencing reads generated in this study have been deposited in the NCBI Sequence Raw Archive (SRA) under accession number SRR31808354, and the corresponding genome assembly is available under BioSample accession number SAMN45949000. BioProject number PRJNA1202041.
FundingThis research was supported by grants from Universidad de Buenos Aires (UBACYT 20020220300083BA) and ANPCYT (Préstamo BID PICT-2020-SERIE A-03132) to Marta Mollerach.
Conflict of interestsNone declared.