After renal transplant, surgical, infection complications, as well as graft rejection may occur; early detection through non-invasive markers is the key to change therapy and avoid biopsy.
ObjectiveThe aim of the study is to determine urine protein profiles in patients undergoing renal transplant with complications and detect its variation when therapy is modified.
Material and methodsUrine samples were collected from patients prior the transplant and various postoperative stages. Urinary protein profiles were obtained by peptide labelling using isobaric isotopes for relative quantification (iTRAQ®).
ResultsA total of 22 patients were included, of whom 12 developed post-transplant complication: 2 with graft rejection (1 male and 1 female) and 10 (6 males and 4 females) in the group of post-transplant infections. Using iTRAQ® 15/345 and 28/113 proteins were identified and fulfilled the acceptance criteria, in graft rejection and post-transplant infections group, respectively.
ConclusionsAlbumin was the only protein found in both groups, the remaining proteins were different. The five proteins with higher scores in graft rejection were: alpha-1-microglobulin, 5′-nucleotidase cytosolic III, retinol-binding protein 4, membrane protein palmitoylated 4, and serine carboxypeptidase, while post-transplant infections were: mitochondrial acetyl-coenzyme A synthetase, putative adenosyl homocysteinase 2, zinc finger protein GLIS1, putative protein FAM157B, and zinc finger protein 615. It remains to elucidate the involvement of each of these in patients with renal transplantation.
En el trasplante renal pueden presentarse complicaciones quirúrgicas, infecciosas y rechazo al injerto; su detección oportuna a través de marcadores no invasivos es la clave para modificar la terapia evitando la biopsia.
ObjetivoDeterminar los perfiles de expresión de proteínas urinarias en pacientes con trasplante renal que desarrollaron complicaciones y detectar su variación al modificar la terapia.
Material y métodosSe recolectaron muestras de orina pretrasplante y de diversas fases del postrasplante; el análisis fue realizado por marcaje peptídico mediante isótopos isobáricos para la cuantificación relativa (iTRAQ®).
ResultadosSe incluyeron 22 pacientes, de los cuales 12 presentaron complicaciones en el postrasplante renal: 2 con rechazo al injerto (un hombre y una mujer) y 10 (6 hombres y 4 mujeres) en el grupo de infecciones sistémicas. A través de iTRAQ® se identificaron en el rechazo al injerto 15/345 proteínas, y en pacientes con infección, 28/113 proteínas, que cumplieron los criterios de aceptación.
ConclusionesLa albúmina fue la única proteína encontrada en ambos grupos; el resto de las proteínas fueron diferentes. Las 5 proteínas con mayor score en rechazo al injerto fueron alfa-1-microglobulina, 5′-nucleotidasa citosólica, proteína 4 de unión a retinol, proteína de membrana 4 palmitolada y serín carboxipeptidasa, mientras que en el grupo de infecciones fueron la acetil coenzima A sintetasa mitocondrial, adenosil homocisteinasa 2, proteína de dedo de cinc GLIS1, proteína putativa de la isoforma FAM157B y proteína de dedo de cinc 615. Queda por dilucidar la participación de cada una de estas en los pacientes con trasplante renal.
Renal transplant is the best available option for the treatment of chronic end-stage renal disease, and compared to chronic dialysis, it improves quality of life and reduces mortality for most patients. The shortage of organ donations is this therapy's greatest limitation, and therefore, extending the useful life of renal grafts is the main priority1,2.
Analysis by protocol biopsy of the transplanted organ is the method of choice to calculate and predict the risk of graft failure, providing indications as to the possible pathogenic mechanism of its dysfunction, and also serving as a guide to the immunosuppressive therapy to be given3. There are different aetiological mechanisms, such as calcineurin inhibitor toxicity, cellular and humoral rejection, and infections, which cause an initial temporary tissue lesion with the final and common morphology of interstitial fibrosis and tubular atrophy4. For this reason, new diagnostic approaches are currently being formulated to detect and monitor the development of graft dysfunction, and its various histological subtypes, in order to optimise the information currently supplied by histology, with the ultimate goal of creating a non-invasive tool to substitute renal graft biopsy5,6.
Proteomics have been widely applied in the search for markers for the diagnosis and prognosis of various disorders. These diseases include, cardiovascular disease, diseases of the male and female reproductive system, liver and kidney disease, and dialysis-related peritonitis, amongst others7–10. Other quantitative alternatives with great potential are being developed, such as peptide marking by isobaric isotopes for relative quantification (iTRAQ®), which is playing a key role in quantitative differential expression proteomics11.
In this study we determined the profile of urinary proteins associated with renal transplant complications and assessed their modification with specific therapy.
Materials and methodsStudy populationThis was an observational, longitudinal, analytical and prospective study, which included patients who had undergone renal transplant from a cadaveric donor in the Hospital Universitario Dr. José Eleuterio González, Universidad Autónoma de Nuevo León, during the period between 28th January 2009 and 30th May 2013. This protocol was approved by our hospital's Ethical Committee and has been registered with the number HI09-003.
Patient inclusion criteriaPatients over 18 years of age were included who had attended the Transplant Department of the Hospital Universitario Dr. José Eleuterio González and who decided to take part in the protocol, and signed the informed consent form. These were classified into two groups: (1) group of renal transplant patients with acute rejection, including patients presenting with raised creatinine and rejection confirmed by biopsy, and (2) group of systemic infections with renal transplant, with patients presenting infections demonstrated by blood culture.
Patient exclusion criteriaPatients under 18 years of age who do not meet the inclusion criteria.
Biological samplingThe patients presenting acute rejection were monitored in the pre-transplant stages, at 24, 48 and 72h, and at days 7, 15 and 30 post-transplant; blood samples were taken, one of 5ml for serum and another of 3ml for blood with EDTA, using the conventional techniques. The serum was stored in 1ml aliquots at −20°C until use.
The conventional mid-stream technique was used for the urine samples, in patients with permanent urinary catheters the sample was taken by aspirating using a needle through the cone of the catheter; they were stored in six 4ml aliquots at −70°C until use. These samples were gathered in the pre-transplant stages and every third day up to 1 month post-transplant, after which time if the patient did not present complications, they were discarded. If the patients presented with a complication, urine samples were collected daily for the next 4 days, and subsequently every third day until 30 days thereafter; while blood samples were taken daily until the seventh day, and then every week for the next 30 days.
Determination of biochemical and haematological parametersThe biochemical tests of glucose (mg/dl), nitrogen from urea (mg/dl), creatinine (mg/dl), chlorine (mmol/l), sodium (mmol/l), potassium (mmol/l), calcium (mg/dl), phosphorous (mg/dl) and magnesium (mg/dl) were performed using Vitros® DT60 II Ortho Chemistry Clinical Diagnostic (DTSC II, Johnson & Johnson Co., Rochester, USA). The Cell Dyn 1700 (Abbott Co., IL, USA) was used for blood cytometry, i.e., haemoglobin (g/dl) and leukocytes (K/μl) in the blood samples with EDTA.
Urine analysis by iTRAQ®The urine samples were analysed in pools according to the group to which the patients had been assigned (graft rejection and infections’ group), 2ml were taken from each, from which 4ml aliquots were made for each group and each phase of the study: pre-renal transplant phase (PH1), post-renal transplant phase prior to complication (PH2), post-renal transplant phase with complication in course (PH3) and post-renal transplant phase with post-treatment complication (PH4) for subsequent concentration, precipitation, labelling and analysis.
- a.
Concentration of urine samples: an aliquot with 4ml of urine was used and placed in an Ultracel centricon 10kDa, Millipore filter; it was centrifuged at 9500rpm at 20°C until reaching a volume of 200μl. One microlitre of Amresco protease inhibitor cocktail was added and the sample was homogenised. In order to precipitate the proteins, 2.4ml of cold acetone at −20°C were added and they were left to precipitate at −20°C for 12–16h. The samples were centrifuged at 3500rpm for 15min. The supernatant was eliminated and 200μl of Milli-Q® water added.
- b.
Quantification of proteins: a calibration curve was prepared with standards of 10, 20, 30, 40 and 50μg of bovine albumin. A Bio-Rad protein quantification kit, based on Bradford's assay was used. For the reduction, alkylation, digestion, desalination and lyophilisation of proteins, their disulphide bridges were reduced with dithiothreitol 10mM for 30min at 56°C. The samples were then incubated with iodoacetamide 50mM for 30min at ambient temperature protected from the light to enable the alkylation of the cysteins. Finally, trypsin (Promega, Fitchburg, WI, USA) was added in a proportion of 1:50 (enzyme:substrate). The enzymatic digestion was incubated for 18h at 37°C. Finally, the samples were desalinated using Sep-Pack® C18 Vac cartridges (Waters) and lyophilised using a Savant SpeedVac® (Thermo Fisher, San José, CA, USA).
- c.
Labelling with the iTRAQ® kit: the reagent vials with isobaric markers 114, 115, 116 and 117 were left at ambient temperature, the reagent was resuspended, 70μl of ethanol were added to each of the kit's reagents and shaken for 1min. An isobaric marker was added at each study phase, phase PH1 was marked with reagent 114, PH2, with 115, PH3, with marker 116, and PH4 with marker 117. The samples and the reagents were mixed and incubated for 1h at ambient temperature. Finally all the samples were mixed. The liquid chromatography–mass spectrometry system comprised an Accela micro-flow liquid chromatograph (Thermo Fischer Co., San José, CA, USA) with splitter (1/20) and an Q-Orbitrap Velos™ mass spectrometer (Thermo-Fischer Co., San José, CA, USA) with a nano-electrospray ionisation system. The spectrometer was calibrated with a solution (Calmix®) of 10 calibrating molecules, which enabled determinations with accuracies better than 5parts/million. A gradient system of 10–100% B solvent (acetonitrile/acetic acid at 0.1%) was used in the liquid chromatography for 120min on a capillary column, PicoFrit® ProteoPep™ II C18 75μm ID×50mm (New Objective, Inc., Woburn, MA, USA). The flow from the liquid chromatography system was 400nl/min. Collision-induced dissociation and high energy collision dissociation methods were used to fragment the peptides. All the spectros were acquired in positive detection mode. The spectrometric data were analysed against the human database using Proteome Discoverer™ 1.3 software, as SEQUEST platform. The restrictive parameters were set, such as error tolerance for the precursor ions and the daughter ions; and modifications such as: carbamidomethyl cysteine, iTRAQ® labelling on the terminal end and lysines (constant), and oxidation of methionine (variable).
The biochemical and haematological results were averaged±standard deviation, and were analysed using one-factor ANOVA tests. SPSS® version 16 software was used for this. P<0.05.was considered statistically significant.
ResultsIn the study period 48 patients had undergone renal transplant from a cadaveric donor, of whom 22 fulfilled the inclusion criteria and agreed to participate in the protocol after signing the informed consent form.
Ten women (45.5%) and 12 men (54.5%) were included in the study, with an average age of 45±15 years of age. Of the 22 patients included, 12 (54.5%) presented complications in the post-renal transplant phase with an average age of 53±17 years of age; 10 (45.5%) had infections, with an average age of 43±17 years of age, and 2 (9%) had acute rejection and both patients were aged 63.
The aetiology of the renal disease of the patients with acute rejection was diabetic nephropathy in both cases (100%), whereas in the systemic infection group the aetiologies were: 2 (20%) patients with membranous glomerulonephritis, 2 (20%) with glomeruloesclerosis, 2 (20%) with diabetic nephropathy, 1 (10%) with severe lupus nephropathy and 2 (20%) of unknown aetiology.
Of the transplanted patients with infections (n=10), 6 presented a surgical complication other than infection: one case with Staphylococcus aureus with arterial pseudoaneurysm; one with Enterococcus faecalis and surgical wound infection; one with Staphylococcus sp. and arteriovenous fistula; one with E. faecalis and fibrosis of the ureter; one with Pseudomonas aeruginosa infection with urinary fistula, and another case with Escherichia coli with urinary fistula; the four remaining cases evolved with no complications and the following aetiological agents were identified: E. coli, Clostridium difficile, E. faecalis and S. aureus.
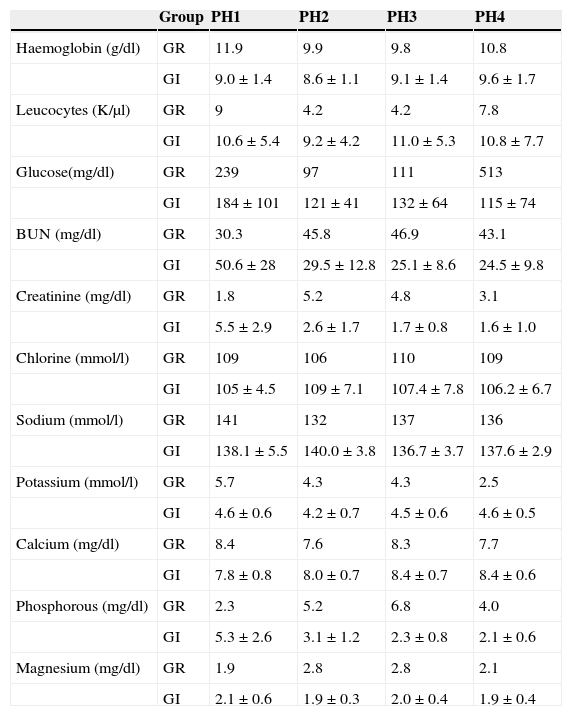
The results of the biochemical and haematological parameters of each group of complications in the different phases of the study are shown in Table 1. In the case of the graft rejection group only the averages of the values are shown, as there were only two cases it was not possible to obtain the standard deviation.
Biochemical and haematological parameters of the groups in the four study phases.
| Group | PH1 | PH2 | PH3 | PH4 | |
|---|---|---|---|---|---|
| Haemoglobin (g/dl) | GR | 11.9 | 9.9 | 9.8 | 10.8 |
| GI | 9.0±1.4 | 8.6±1.1 | 9.1±1.4 | 9.6±1.7 | |
| Leucocytes (K/μl) | GR | 9 | 4.2 | 4.2 | 7.8 |
| GI | 10.6±5.4 | 9.2±4.2 | 11.0±5.3 | 10.8±7.7 | |
| Glucose(mg/dl) | GR | 239 | 97 | 111 | 513 |
| GI | 184±101 | 121±41 | 132±64 | 115±74 | |
| BUN (mg/dl) | GR | 30.3 | 45.8 | 46.9 | 43.1 |
| GI | 50.6±28 | 29.5±12.8 | 25.1±8.6 | 24.5±9.8 | |
| Creatinine (mg/dl) | GR | 1.8 | 5.2 | 4.8 | 3.1 |
| GI | 5.5±2.9 | 2.6±1.7 | 1.7±0.8 | 1.6±1.0 | |
| Chlorine (mmol/l) | GR | 109 | 106 | 110 | 109 |
| GI | 105±4.5 | 109±7.1 | 107.4±7.8 | 106.2±6.7 | |
| Sodium (mmol/l) | GR | 141 | 132 | 137 | 136 |
| GI | 138.1±5.5 | 140.0±3.8 | 136.7±3.7 | 137.6±2.9 | |
| Potassium (mmol/l) | GR | 5.7 | 4.3 | 4.3 | 2.5 |
| GI | 4.6±0.6 | 4.2±0.7 | 4.5±0.6 | 4.6±0.5 | |
| Calcium (mg/dl) | GR | 8.4 | 7.6 | 8.3 | 7.7 |
| GI | 7.8±0.8 | 8.0±0.7 | 8.4±0.7 | 8.4±0.6 | |
| Phosphorous (mg/dl) | GR | 2.3 | 5.2 | 6.8 | 4.0 |
| GI | 5.3±2.6 | 3.1±1.2 | 2.3±0.8 | 2.1±0.6 | |
| Magnesium (mg/dl) | GR | 1.9 | 2.8 | 2.8 | 2.1 |
| GI | 2.1±0.6 | 1.9±0.3 | 2.0±0.4 | 1.9±0.4 |
PH1, pre-renal transplant phase; PH2, post-renal transplant phase prior to complication; PH3, post-renal transplant phase complication in course; PH4, post-renal transplant phase complication treated; GI, group of patients with infection; GR, group of patients with graft rejection.
The biochemical and haematological parameters were compared between the phases for the infections’ group. The statistically significant values were: glucose PH1 (184±101mg/dl) vs. PH2 (121±41mg/dl), P=0.009; nitrogen from urea PH1 (50.6±28mg/dl) vs. PH3 (25.1±8.6mg/dl), P=0.018, and PH1 (50.6±28mg/dl) vs. PH4 (24.5±9.8mg/dl), P=0.049; creatinine PH1 (5.5±2.9mg/dl) vs. PH3 (1.7±0.8mg/dl), P=0.004, PH1 (5.5±2.9mg/dl) vs. PH4 (1.6±1.0mg/dl), P=0.013, and PH2 (2.6±1.7mg/dl) vs. PH3 (1.7±0.8mg/dl), P=0.020; and magnesium in PH1 (2.1±0.6mg/dl) vs. PH2 (1.9±0.3mg/dl), P=0.050.
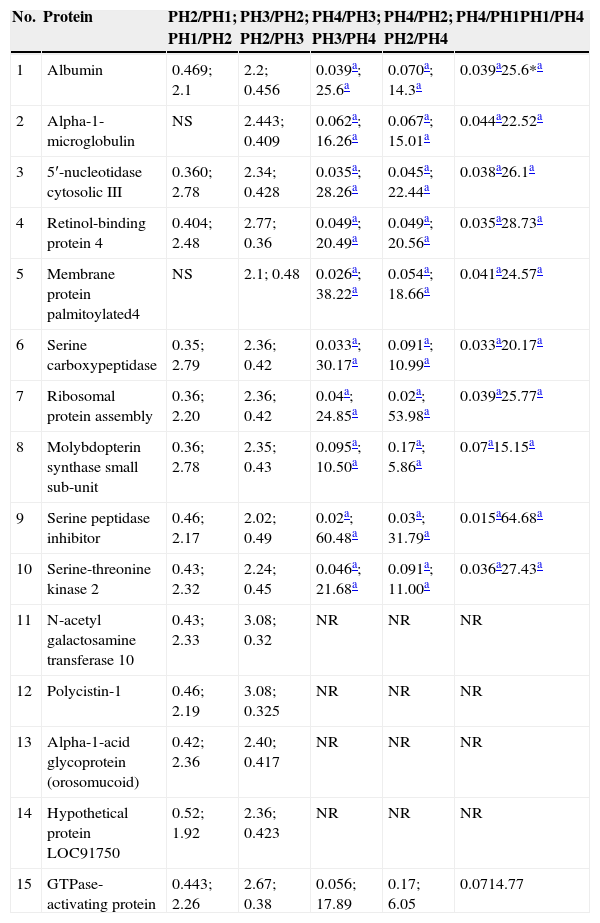
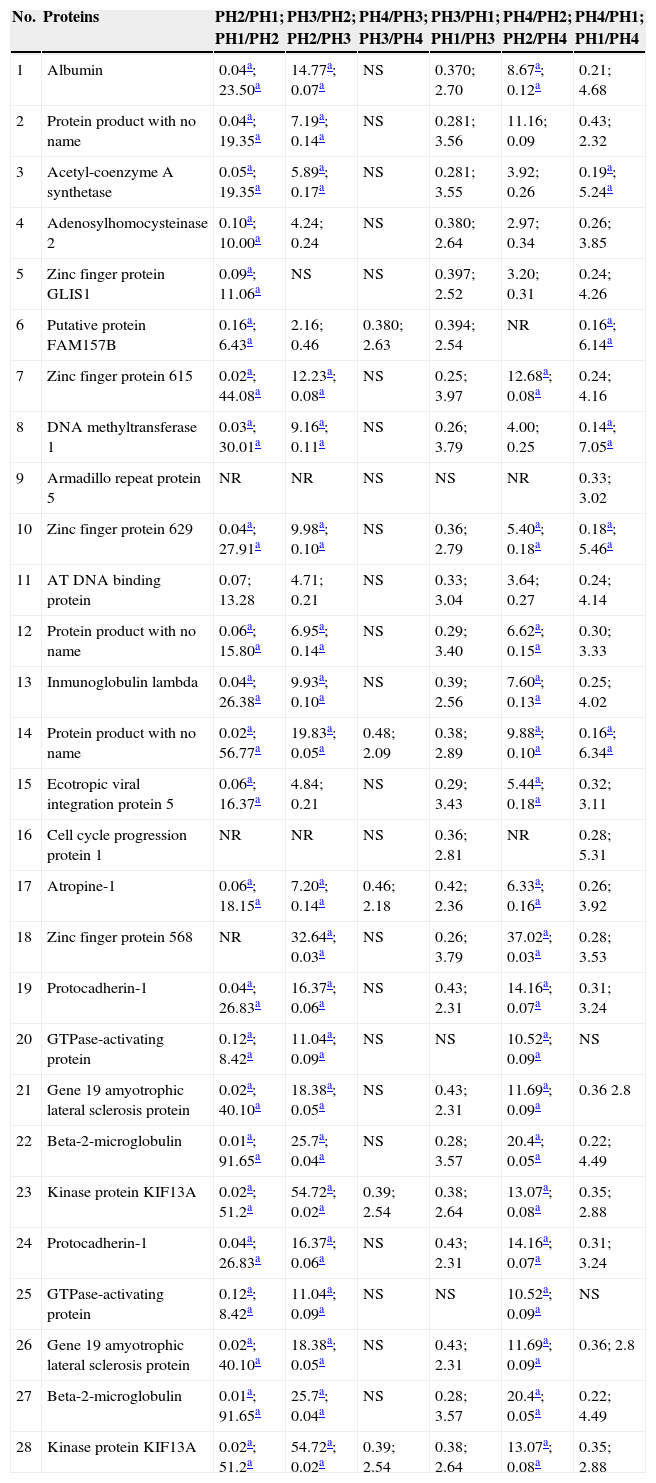
With the application of iTRAQ® techniques using liquid chromatography–mass spectrometry system, 345 proteins were found in the graft rejection group, of which 15 met the acceptance criteria for the technique (score>30, 2 or more peptides identified with 95% confidence) and all of them were human peptides (Table 2), whereas in the group of patients with infections, 113 proteins were found, of which 28 fulfilled the acceptance criteria for the technique (Table 3). Differences with twice the abundance of proteins between the study phases were considered significant.
Proteins in the group of patients with graft rejection who fulfilled the acceptance criteria.
| No. | Protein | PH2/PH1; PH1/PH2 | PH3/PH2; PH2/PH3 | PH4/PH3; PH3/PH4 | PH4/PH2; PH2/PH4 | PH4/PH1PH1/PH4 |
|---|---|---|---|---|---|---|
| 1 | Albumin | 0.469; 2.1 | 2.2; 0.456 | 0.039a; 25.6a | 0.070a; 14.3a | 0.039a25.6*a |
| 2 | Alpha-1-microglobulin | NS | 2.443; 0.409 | 0.062a; 16.26a | 0.067a; 15.01a | 0.044a22.52a |
| 3 | 5′-nucleotidase cytosolic III | 0.360; 2.78 | 2.34; 0.428 | 0.035a; 28.26a | 0.045a; 22.44a | 0.038a26.1a |
| 4 | Retinol-binding protein 4 | 0.404; 2.48 | 2.77; 0.36 | 0.049a; 20.49a | 0.049a; 20.56a | 0.035a28.73a |
| 5 | Membrane protein palmitoylated4 | NS | 2.1; 0.48 | 0.026a; 38.22a | 0.054a; 18.66a | 0.041a24.57a |
| 6 | Serine carboxypeptidase | 0.35; 2.79 | 2.36; 0.42 | 0.033a; 30.17a | 0.091a; 10.99a | 0.033a20.17a |
| 7 | Ribosomal protein assembly | 0.36; 2.20 | 2.36; 0.42 | 0.04a; 24.85a | 0.02a; 53.98a | 0.039a25.77a |
| 8 | Molybdopterin synthase small sub-unit | 0.36; 2.78 | 2.35; 0.43 | 0.095a; 10.50a | 0.17a; 5.86a | 0.07a15.15a |
| 9 | Serine peptidase inhibitor | 0.46; 2.17 | 2.02; 0.49 | 0.02a; 60.48a | 0.03a; 31.79a | 0.015a64.68a |
| 10 | Serine-threonine kinase 2 | 0.43; 2.32 | 2.24; 0.45 | 0.046a; 21.68a | 0.091a; 11.00a | 0.036a27.43a |
| 11 | N-acetyl galactosamine transferase 10 | 0.43; 2.33 | 3.08; 0.32 | NR | NR | NR |
| 12 | Polycistin-1 | 0.46; 2.19 | 3.08; 0.325 | NR | NR | NR |
| 13 | Alpha-1-acid glycoprotein (orosomucoid) | 0.42; 2.36 | 2.40; 0.417 | NR | NR | NR |
| 14 | Hypothetical protein LOC91750 | 0.52; 1.92 | 2.36; 0.423 | NR | NR | NR |
| 15 | GTPase-activating protein | 0.443; 2.26 | 2.67; 0.38 | 0.056; 17.89 | 0.17; 6.05 | 0.0714.77 |
PH1, pre-renal transplant phase; PH2, post-renal transplant phase prior to complication; PH3, post-renal transplant phase complication in course; PH4, post-renal transplant phase complication treated; GI, group of patients with infection; GR, group of patients with graft rejection; NT, not taken; NS, not significant.
Proteins in the group of patients with infections which fulfil the acceptance criteria.
| No. | Proteins | PH2/PH1; PH1/PH2 | PH3/PH2; PH2/PH3 | PH4/PH3; PH3/PH4 | PH3/PH1; PH1/PH3 | PH4/PH2; PH2/PH4 | PH4/PH1; PH1/PH4 |
|---|---|---|---|---|---|---|---|
| 1 | Albumin | 0.04a; 23.50a | 14.77a; 0.07a | NS | 0.370; 2.70 | 8.67a; 0.12a | 0.21; 4.68 |
| 2 | Protein product with no name | 0.04a; 19.35a | 7.19a; 0.14a | NS | 0.281; 3.56 | 11.16; 0.09 | 0.43; 2.32 |
| 3 | Acetyl-coenzyme A synthetase | 0.05a; 19.35a | 5.89a; 0.17a | NS | 0.281; 3.55 | 3.92; 0.26 | 0.19a; 5.24a |
| 4 | Adenosylhomocysteinase 2 | 0.10a; 10.00a | 4.24; 0.24 | NS | 0.380; 2.64 | 2.97; 0.34 | 0.26; 3.85 |
| 5 | Zinc finger protein GLIS1 | 0.09a; 11.06a | NS | NS | 0.397; 2.52 | 3.20; 0.31 | 0.24; 4.26 |
| 6 | Putative protein FAM157B | 0.16a; 6.43a | 2.16; 0.46 | 0.380; 2.63 | 0.394; 2.54 | NR | 0.16a; 6.14a |
| 7 | Zinc finger protein 615 | 0.02a; 44.08a | 12.23a; 0.08a | NS | 0.25; 3.97 | 12.68a; 0.08a | 0.24; 4.16 |
| 8 | DNA methyltransferase 1 | 0.03a; 30.01a | 9.16a; 0.11a | NS | 0.26; 3.79 | 4.00; 0.25 | 0.14a; 7.05a |
| 9 | Armadillo repeat protein 5 | NR | NR | NS | NS | NR | 0.33; 3.02 |
| 10 | Zinc finger protein 629 | 0.04a; 27.91a | 9.98a; 0.10a | NS | 0.36; 2.79 | 5.40a; 0.18a | 0.18a; 5.46a |
| 11 | AT DNA binding protein | 0.07; 13.28 | 4.71; 0.21 | NS | 0.33; 3.04 | 3.64; 0.27 | 0.24; 4.14 |
| 12 | Protein product with no name | 0.06a; 15.80a | 6.95a; 0.14a | NS | 0.29; 3.40 | 6.62a; 0.15a | 0.30; 3.33 |
| 13 | Inmunoglobulin lambda | 0.04a; 26.38a | 9.93a; 0.10a | NS | 0.39; 2.56 | 7.60a; 0.13a | 0.25; 4.02 |
| 14 | Protein product with no name | 0.02a; 56.77a | 19.83a; 0.05a | 0.48; 2.09 | 0.38; 2.89 | 9.88a; 0.10a | 0.16a; 6.34a |
| 15 | Ecotropic viral integration protein 5 | 0.06a; 16.37a | 4.84; 0.21 | NS | 0.29; 3.43 | 5.44a; 0.18a | 0.32; 3.11 |
| 16 | Cell cycle progression protein 1 | NR | NR | NS | 0.36; 2.81 | NR | 0.28; 5.31 |
| 17 | Atropine-1 | 0.06a; 18.15a | 7.20a; 0.14a | 0.46; 2.18 | 0.42; 2.36 | 6.33a; 0.16a | 0.26; 3.92 |
| 18 | Zinc finger protein 568 | NR | 32.64a; 0.03a | NS | 0.26; 3.79 | 37.02a; 0.03a | 0.28; 3.53 |
| 19 | Protocadherin-1 | 0.04a; 26.83a | 16.37a; 0.06a | NS | 0.43; 2.31 | 14.16a; 0.07a | 0.31; 3.24 |
| 20 | GTPase-activating protein | 0.12a; 8.42a | 11.04a; 0.09a | NS | NS | 10.52a; 0.09a | NS |
| 21 | Gene 19 amyotrophic lateral sclerosis protein | 0.02a; 40.10a | 18.38a; 0.05a | NS | 0.43; 2.31 | 11.69a; 0.09a | 0.36 2.8 |
| 22 | Beta-2-microglobulin | 0.01a; 91.65a | 25.7a; 0.04a | NS | 0.28; 3.57 | 20.4a; 0.05a | 0.22; 4.49 |
| 23 | Kinase protein KIF13A | 0.02a; 51.2a | 54.72a; 0.02a | 0.39; 2.54 | 0.38; 2.64 | 13.07a; 0.08a | 0.35; 2.88 |
| 24 | Protocadherin-1 | 0.04a; 26.83a | 16.37a; 0.06a | NS | 0.43; 2.31 | 14.16a; 0.07a | 0.31; 3.24 |
| 25 | GTPase-activating protein | 0.12a; 8.42a | 11.04a; 0.09a | NS | NS | 10.52a; 0.09a | NS |
| 26 | Gene 19 amyotrophic lateral sclerosis protein | 0.02a; 40.10a | 18.38a; 0.05a | NS | 0.43; 2.31 | 11.69a; 0.09a | 0.36; 2.8 |
| 27 | Beta-2-microglobulin | 0.01a; 91.65a | 25.7a; 0.04a | NS | 0.28; 3.57 | 20.4a; 0.05a | 0.22; 4.49 |
| 28 | Kinase protein KIF13A | 0.02a; 51.2a | 54.72a; 0.02a | 0.39; 2.54 | 0.38; 2.64 | 13.07a; 0.08a | 0.35; 2.88 |
PH1, pre-renal transplant phase; PH2, post-renal transplant phase prior to complication; PH3, post-renal transplant phase complication in course; PH4, post-renal transplant phase complication treated; GI, group of patients with infection; GR, group of patients with graft rejection; NT, not taken; NS, not significant.
Of the 15 proteins selected for the group presenting with graft rejection, it was found that the serine peptidase inhibitor, membrane protein palmitoylated 4, serine carboxypeptidase, 5′-nucleotidase cytosolic and molybdopterin synthase small sub-unit were the proteins which presented the highest ratios between phases PH3 and PH4, whereas the proteins with the highest score values were: alpha-1-microglobulin, 5′-nucleotidase cytosolic, retinol-binding protein 4, membrane protein palmitoylated 4 and serine carboxypeptidase for this same group.
In the group of patients with infections, of the 28 proteins, those with the highest ratios between phases PH2 and PH3 were: RNA processing protein 12, glypican 4, zinc finger protein 568, kinase protein KIF13A and protocadherin-1; and those with the highest score were mitochondrial acetyl-coenzyme A synthetase, putative adenosyl homocysteinase 2, putative protein FAM157B, zinc finger protein 615 and DNA methyltransferase 1.
DiscussionRenal transplant is the best available option for the treatment of end-stage chronic renal disease, and compared to chronic dialysis, it improves quality of life and reduces mortality for the majority of patients. The shortage of organ donors is this therapy's major limitation, and therefore the main priority is to extend the useful life of renal grafts1,2.
New non-invasive diagnostic approaches are currently being developed to detect and monitor the evolution of graft dysfunction9–11; proteomics have been widely used to look for markers for the diagnosis and prognosis of various diseases10,12–14. Other quantitative alternatives with great potential have been developed recently, such as iTRAQ®, which is playing an important role in quantitative differential expression proteomics. It is predicted that proteomics will play a major part in the area of nephrology in the short term, and that this progress will require interactive dialogue and collaboration between clinical and analytical specialists15,16.
In this study we describe a series of proteins associated with various post renal transplant complications, such as graft rejection and infections, and the modification of these proteins with the therapy specific to each of them. This study is the first in our country to describe the proteins which are altered in renal transplant patients who go on to reject the graft, and in those who present infectious complications.
By labelling using the iTRAQ® technique and final analysis by liquid chromatography–mass spectrometry, a total of 345 proteins were found in the graft rejection group, of which only 15 met the acceptance criteria described above (Table 2). While in the group of patients with infections, 28 of the 113 proteins identified fulfilled the technique's acceptance criteria (Table 3).
Of the 15 proteins found in the graft rejection group, the five presenting the highest ratios between phases PH3 and PH4 were: serine peptidase inhibitor which is activated in response to cellular stress and during renal ischaemia-reperfusion injury17,18, likewise, it has been reported that levels of the inhibitor correlate with the severity of the rejection, it being possible to predict the subsequent function of the renal allograft19,20; membrane protein palmitoylated 4, an integral membrane protein with guanylate kinase activity, which allows it to interact with the cytoskeleton and regulate cell proliferation and signal transduction21; serine carboxypeptidase, a protease of unknown function which was first classified in human macrophages, believed to be limited to a great extent to the monocytic strain, although recent studies demonstrate that it might also be expressed by cells outside the immune system22; 5′-nucleotidase cytosolic, which is involved in various functions, such as cell–cell communication, nucleic acid repair, synthesis of nucleotides, signal transduction and membrane transport23, and finally, molybdopterin synthase small sub-unit, which in humans is involved in the biosynthesis of molybdenum cofactor, a genetic deficiency that results in autosomal recessive disease which is generally fatal with severe neurological symptoms24. The highest ratios between phases PH3 vs. PH4 of these five proteins show that they are useful candidates as biomarkers of poor renal graft function. Moreover, it can be seen that in PH4, with the modification of immunosuppressive therapy, the expression of these five proteins is significantly reduced, demonstrating their usefulness as potential biomarkers of good renal graft function in response to immunosuppressive therapy.
Furthermore, for this same group of patients, the five proteins with the highest score were: alpha-1-microglobulin, which is a lipocalin with immunosuppressive properties25, which eliminates free radicals26, and is one of the proteins that has been most studied as a potential non-invasive biomarker for the early detection of kidney abnormalities, as well as for its usefulness in differentiating nephrological and urological diseases27; 5′-nucleotidase cytosolic, whose pharmacological inhibition has been reported to increase CAM concentrations and improve renal function in rat experimental models of ischaemia and reperfusion28; retinol-binding protein 4, which has been reported as raised in patients with interstitial tubular injury29 and is secreted by the adipocytes, and has been reported as raised in cases of insulin resistance30; membrane protein palmitoylated 4, which has been seen over-expressed in renal tissue subjected to ischaemia-reperfusion processes31, and serine carboxypeptidase, which is particularly expressed in the heart and kidneys22. Similar to several publications on renal function, we found in this study that these five proteins are over-expressed during the episode of acute graft rejection and their expression is significantly low when the patients are not presenting renal graft rejection or are responding to the modification made to their immunosuppressive therapy.
In the group of patients with infections, of the 28 proteins, those which presented the highest ratios between phases PH2 vs. PH3 were: RNA processing protein 12; glypican 4, of the heparan sulphate proteoglycans family of the cell surface which express in a regulated manner by developing a specific tissue32; zinc finger protein 568, which has been described as having a protective effect on various stress conditions, such as thermal shock and osmotic shock33; kinase protein KIF13A, which are proteins of the kinase superfamily, that are protagonists in the intracellular transport system, which is essential for cell function and morphology34; and protocadherin-1 protein, a protein family which it has been suggested plays a role in brain development, glomerular cleft formation, and can also act as tumour suppressors, although their mechanisms of action have not been explained35.
The proteins with the highest score for this group with infections were: isoform 3 of mitochondrial acetyl-coenzyme A synthetase, described as important in maintaining normal body temperature during fasting, and energy homeostasis36; putative adenosyl homocysteinase 2, whose expression increases considerably during activation of blood and skin cells37; putative protein FAM157B, whose function has not been reported; zinc finger protein 615, which participates in transcription regulation38, and DNA methyltransferase 1, which participates in the regulation of gene expression according to a methylation pattern39.
Furthermore, massive proteinuria has been recognised as a risk factor for graft failure in the long term40. A recent study indicated that low-grade proteinuria was a strong predictor, irrespective of graft failure long term41. In this study, albumin was expressed in the two complications evaluated, therefore it was not useful as a specific biomarker of any of the complications studied, but it is an indicator of renal damage, as it is one of the proteins which are most expressed prior to transplant; therefore when the transplant is performed, its expression diminishes considerably, but if any type of complication presents, it over-expresses again.
As described in this study, various proteins are candidates as biomarkers of good renal graft function, since their presence in the urine can be used to distinguish the patients who have good graft function from those who do not. Furthermore, each of the complications assessed presented a series of different proteins, apart from albumin. This study emphasizes the usefulness of urinary proteomics in identifying biomarkers and in the future design of a non-invasive diagnostic tool to better detect transplanted renal graft dysfunction.
Conflict of interestsThe authors have no conflict of interests to declare.
Please cite this article as: Escobedo-Villarreal MM, Mercado-Moreira AB, Muñoz-Espinosa LE, Gamboa-Esparza M, Pérez-Rodríguez E, Cordero-Pérez P. Detección de proteínas urinarias por iTRAQ® asociadas a complicaciones del trasplante renal y su modificación con la terapia. Cir Cir. 2015;83:393–401.