Sour cassava starch (Polvilho azedo) is obtained from a spontaneous fermentation conducted by microorganisms from raw materials and fermentation tanks. This product is traditionally used in the baking industry for the manufacture of biscuits and Brazilian cheese breads. However, the end of fermentation is evaluated empirically, and the process occurs without standardization, which results in products of inconsistent quality. Predominant microbiota from a cassava flour manufacturer was isolated in order to select starter cultures for the production of sour cassava starch in a pilot-scale fermentation process. Lactic acid bacteria and yeasts were isolated, enumerated and grouped by Restriction Fragment Length Polymorphism, and PCR fingerprinting, respectively. One isolate of each molecular profile was identified by sequencing of the rRNA gene. LAB were prevalent throughout the entire process. Lactobacillus brevis (21.5%), which produced the highest values of acidity, and Lactobacillus plantarum (13.9%) were among the most frequent species. Pichia scutulata (52.2%) was the prevalent yeast and showed amylolytic activity. The aforementioned species were tested as single and mixed starter cultures in a pilot-scale fermentation process for 28 days. L. plantarum exhibited better performance as a starter culture, which suggests its potential for the production of sour cassava starch.
Because cassava (Manihot esculenta CRANTZ) has a high starch content (approximately 80%), it is an important source of carbohydrates that can be sold fresh or processed into a variety of value-added products; cassava is characterized as a multipurpose crop.1 In 2014, the estimated Brazilian cassava production was 23 million tons.2 Although widely consumed, cassava has limitations due to its perishability, toxicity and low protein content.3 The root of cassava contains cyanogenic glycosides that act as defence substances through the release of hydrogen cyanide, which is responsible for its toxicity. The traditional fermentations of cassava are quite suitable for the detoxification, preservation and development of products with desirable viscoelastic texture.4 Lactic fermentation not only extends the shelf life of this root but also decreases its toxicity.5
Natural fermentation of wet starch extracted from cassava root is a traditional technology widely used in Latin America. Sweet cassava starch and sour cassava starch undergo the same process of starch extraction, but differ in fermentation time (from two to seven days and from 20 to 70 days, respectively) and therefore have different levels of acidity, maximum of 1% of acidity and 5% of acidity, respectively. Sour cassava starch stands out from sweet cassava starch and other flour in the preparation of bakery products for its unique expansion capacity, without the addition of baking soda and in the absence of gluten. Sour cassava starch is a typical Brazilian food generally produced by small and medium-sized rural industries.6 As sour cassava starch is essentially handcrafted, even though it is produced in modern cassava flour manufacturers, it still has heterogeneous physicochemical and sensory quality. The succession of microorganisms from raw materials and fermentation tanks occurs naturally during cassava fermentation and results in a microbiota with a prevalence of lactic acid bacteria (LAB), such as Lactobacillus plantarum, Lactobacillus fermentum, Lactobacillus brevis and Leuconostoc mesenteroides.7–10 In the production of sour cassava starch in Brazil, Lactobacillus occurs in association with yeasts, such as Galactomyces geothricum and Issatchenkia (now Pichia) sp.6,11
The wide range and complexity of cassava spontaneous fermentation microbiota are the main factors responsible for the lack of homogeneity and low product quality. The use of selected strains is an important alternative because it provides less variation in the content of chemical compounds, shorter fermentation time, higher yield and sensorial quality.12,13 The LAB, which are used as natural or selected starter cultures in fermented foods, are able to acidify and enhance the flavor. Furthermore, the LAB can protect food from the development of pathogens due to the formation of antimicrobial compounds.14,15
Although sour cassava starch is widely consumed in Latin America and gained prominence in the preparation of bakery products with a low level or the absence of gluten, there are few studies on its fermentation process. Therefore, the study of starter cultures contributes significantly to the understanding and optimization of sour cassava starch production. The present work aimed to select the starter cultures with the appropriate characteristics for the production of sour cassava starch in a pilot-scale fermentation process.
Materials and methodsCollection of samples from cassava flour manufacturerA total of 16 samples of 100g each were collected from a cassava flour manufacturer in the municipality of Formiga, Minas Gerais (MG) state, Brazil, on days 0, 5, 12, 19, 26, 33 and 40 of a 56-day spontaneous fermentation from a fermentation tank with usable capacity of 16,000L. These samples were transported to the laboratories of Food Microbiology and Industrial Microbiology and Biocatalysis (Faculdade de Farmácia, Universidade Federal de Minas Gerais, MG, Brazil) on ice and processed within 24h. The processing consisted of weighing twenty-five grams of each sample in sterile flasks, diluted in 225mL of 1gL−1 peptone water, and preparing serial decimal dilutions.16
Identification of lactic acid bacteria found on samples from cassava flour manufacturerAppropriate decimal dilutions were spread on de Man, Rogosa and Sharpe (MRS; Acumedia, Lansing, MI, USA) agar containing 0.1gL−1 cycloheximide16 and incubated in anaerobic jars of 2.5L (Permution, Curitiba, Brazil) at 37°C for 48h. After growth, one colony of each morphotype was counted and purified for later identification. Each isolate was Gram stained and then subjected to the catalase test.17
DNA was extracted with an adaptation of the method described by Hoffman and Winston.18 The colonies previously grown on MRS agar were resuspended in 100μL of Tris–EDTA (TE). Then, 100μL of phenol–chloroform–isoamyl alcohol (25:24:1) and 0.3g of glass beads were added to the suspension. Tubes containing this mixture were homogenized by a vortex shaker (QL-901, Biomixer, Santa Clara, CA, USA) for three to 4min and centrifuged at 13,000rpm for 5min (Eppendorf, Hamburg, Hamburg, Germany). After that, the supernatant was transferred to another tube. Then, a volume of 960mLL−1 ethanol corresponding to the volume of the supernatant recovered was added to the tube. The tubes were homogenized by inversion and centrifuged at 13,000rpm for 2min. The liquid phase was discarded, the tubes were dried overnight, and the DNA resuspended in 50μL of TE. The DNA concentration was determined by NanoDrop ND 1000 Spectrophotometer (NanoDrop Products, Wilmington, DE, USA). The DNA of lactic acid bacteria was subjected to PCR amplification of the 16S rRNA gene using the primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′).19 All LAB isolates were grouped by Restriction Fragment Length Polymorphism (RFLP) by digestion with restriction enzymes MspI, HinfI and HaeIII (Promega Corporation, Madison, WI, USA) according to the modified methodology of Brightwell et al.20 For the digestion reaction, 2μL of 10× buffer, 2μL of bovine serum albumin (BSA) only for the MspI enzyme, 1μL of enzyme, DNA ≤1500ng/μL and water q.s.p. 20μL. The tubes were incubated at 37°C for 3h. The restriction fragments obtained were separated by 2% agarose gel electrophoresis (Pronadisa, Spain) in 0.5% TBE buffer at 100V. The gels were stained with GelRedTM solution (Biotium, USA) and visualized under ultraviolet light (UV) by an image capture system (Vilber Lourmat, France). An isolate of each different molecular RFLP profile was selected and subjected to 16S rRNA sequence analysis.19 Samples were sequenced by capillary electrophoresis in ABI3130 equipment using POP7 polymer and BigDye v3.1 (Myleus Biotechnology, Belo Horizonte, MG, Brazil). The sequences were assembled, edited and aligned with the program MEGA6.21 The sequences obtained were compared with those included in the GenBank database using the Basic Local Alignment Search Tool (BLAST at http://www.ncbi.nlm.nih.gov).
Differentiation of species of the L. plantarum groupAll the isolates that were presumably identified by 16S rRNA sequence analysis as belonging to the L. plantarum group were subjected to Multiplex PCR Assay with recA Gene-Derived Primers. This analysis allows the separation of the three closely related species from the L. plantarum group by comparison of the size of their amplicons: 318 bp for L. plantarum, 218 bp for Lactobacillus pentosus and 107 bp for Lactobacillus paraplantarum.22L. plantarum ATCC1 4917, L. paraplantarum DSM 10667 and L. pentosus ATCC 8041 were used as controls.
Identification of yeasts found on samples from cassava flour manufacturerPortions of appropriate decimal dilutions were spread onto yeast extract–malt extract (YM; Acumedia, Lansing, MI, USA) agar containing 0.2gL−1 chloramphenicol and incubated for 48h at 25°C under aerobic conditions.6 One colony of each different morphotype was counted and purified for later identification.
The total DNA of each yeast isolate was extracted with an adaptation of the method described by da Silva-Filho et al.23 The colonies previously grown on YM agar were resuspended in 100μL of lysis buffer (Tris–HCl – trishydroxymethylaminomethane – 0.05M, 0.05M EDTA, 0.1M NaCl and 10gL−1 SDS – sodium dodecyl sulfate). The tubes containing the suspension were incubated in a water bath at 65°C for 35min. Then, 100μL of phenol–chloroform–isoamyl alcohol (25:24:1) were added to the suspension. Tubes containing this mixture were homogenized by a vortex shaker (QL-901, Biomixer, Santa Clara, CA, USA) for 3min and centrifuged for 15min (Eppendorf, Hamburg, Hamburg, Germany). After that, the supernatant was transferred to another tube, to which was added 100μL of cold 700mLL−1 ethanol. The tubes were homogenized by inversion and centrifuged at 13,000rpm for 3min (Eppendorf, Hamburg, Hamburg, Germany). The liquid phase was discarded and the tubes were dried overnight. The DNA was resuspended in 100μL of TE, and stored in a freezer at −20°C. The DNA concentration was determined by NanoDrop ND 1000 Spectrophotometer (NanoDrop Products, Wilmington, DE, USA).
All yeast isolates were grouped by their molecular profiles using the PCR fingerprinting technique with the microsatellite primer (GTG)5 (5′-GTGGTGGTGGTGGTG-3′).24 The PCR mixture contained 2.5μL of 10× buffer, 1μL of dNTPs 0.1mM, 1.5μL of MgCl2, 2μL of the primer (GTG)5 10μmol−1 (Invitrogen, Carlsbad, CA, USA), 0.2μL of Taq DNA polymerase 1U and 1μL of DNA in a total volume of 25μL. The PCR reaction was performed on the Mastercycler® Pro and showed the following conditions: initial denaturation at 94°C for 2min, 40 cycles of denaturation at 95°C for 45s, annealing at 50°C for 1min and extension at 72°C for 1min, followed by final extension at 72°C for 6min. One representative of each different molecular profile was identified by sequencing of the D1/D2 domains of the large subunit of rRNA gene using the primers NL1 (5′-GCATATCAATAAGCGGAGGAAAAG-3′) and NL4 (5′-GGTCCGTGTTTCAAGACGG-3′) according to Lachance et al.25 The PCR mixture contained 5μL of 10× buffer, 1μL of dNTPs 0.05mM, 3μL of MgCl2 1.5mM, 1μL of the primer NL1 10μmol−1 (Invitrogen, Carlsbad, CA, USA), 1μL of the primer NL4 10μmol−1 (Invitrogen, Carlsbad, CA, USA), 0.2μL of Taq DNA polymerase 1U and 1μL of DNA in a total volume of 50μL. The reaction was performed on the Mastercycler® Pro and showed the following conditions: initial denaturation at 95°C for 2min, 35 cycles of denaturation at 95°C for 15s, annealing at 54°C for 25s, and extension at 72°C for 20s, followed by final extension at 72°C for 10min. The samples were sequenced by capillary electrophoresis in ABI3130 equipment using POP7 polymer and BigDye v3.1. Sequence analyses were performed as described above.
Evaluation of samples from cassava flour manufacturerTotal titratable acidity (TTA) and pH of samples were determined.26 The final product from the fermentation tank was submitted to microbiological analyses to search for Bacillus cereus, thermotolerant coliforms and Salmonella spp. according to Brazilian legislation.27
Screening for selection of strains to be tested as single or mixed starter cultureAll the strains of LAB species isolated were tested in triplicate for starch degradation on plates with MRS agar containing 20gL−1 soluble starch28 and all the strains of yeast species isolated, on plates with YM agar containing 20gL−1 soluble starch.29 After growth, the revelation was performed with iodine solution in order to view starch hydrolysis halos. Total acid production was evaluated in 100mL of broth containing 20gL−1 (non-fermented) cassava starch, 10gL−1 glucose and 5gL−1 of beef extract with 24 and 48h. Aliquots of 10mL of broth were used for TTA measurement.26 The strains that showed starch degradation halos and higher values of TTA were subsequently tested as starter cultures.
Use of starter cultures in a pilot-scale fermentation processSingle cultures of selected LAB strains and mixed cultures of these in association with the yeast were inoculated into 100mL of culture medium containing 20gL−1 (non-fermented) cassava starch, 10gL−1 glucose and 5gL−1 beef extract and then incubated at room temperature for 24–48h. The entire fermented broth was used to inoculate 500mL of the same medium for 24–48h. The resulting broth was used to inoculate 5L of a medium containing 100gL−1 cassava starch, for 28 days. The 5-L fermentations were inoculated with 8log10CFU/mL of LAB and/or 6log10CFU/mL of yeasts. Assays were performed without aeration.
Samples were weekly collected in triplicate for determination of pH and TTA26 as well as for verification of the viability of cultures. The monitoring of starter cultures was done by isolation and purification of three colonies per week of each LAB or yeast that resembled morphologically to the starter cultures selected and performed with molecular methods described above, and conducted by comparison of profiles obtained with profiles already known. After the end of fermentation, the material was dried at room temperature for 5 days, packed and stored under refrigeration until completion of measurements of pH and TTA26; expansion capacity30; and microbiological analysis, as described above. The expansion capacity assay was carried out according to the procedure proposed by Tropical Roots Center. Forty milliliters of boiling distilled water were added to 50g of cassava starch. Each mass was modeled for making round cookies of approximately 10g each. The diameters of the cookies were measured with a universal pachymeter (Series 530, Mitutoyo) before and after taking them to the oven for a period of 20min at 220°C. The expansion capacity was calculated by the ratio of averages of the final diameter (after baking) and the initial diameter (before baking).
Data analysisThe data obtained were subjected to analysis of variance (ANOVA) and significant differences between means were determined by Tukey's test with a 0.05 significance level.
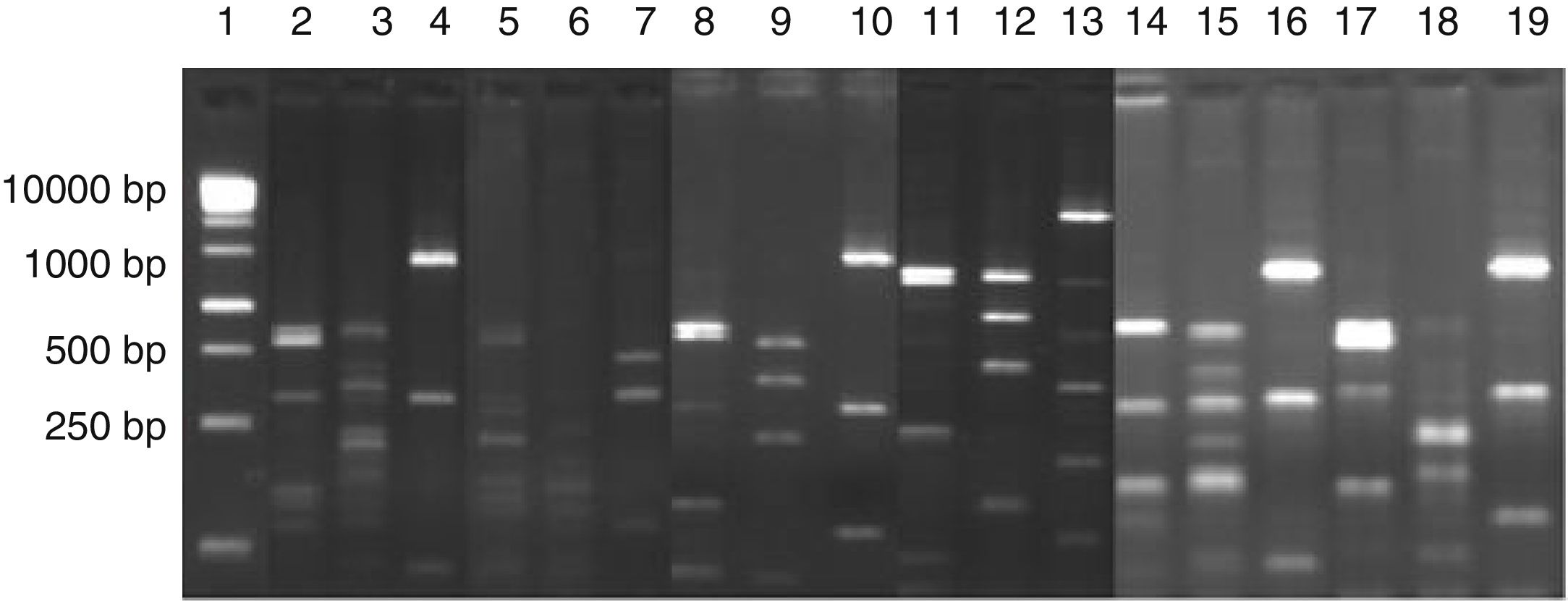
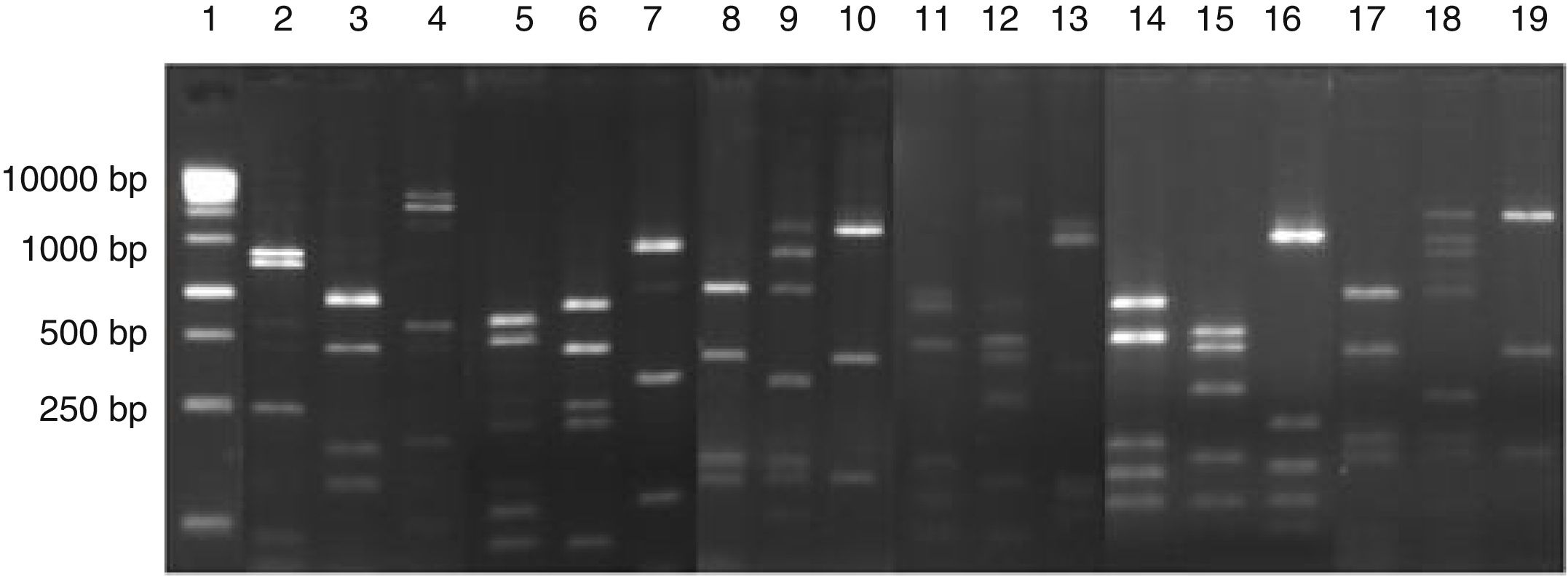
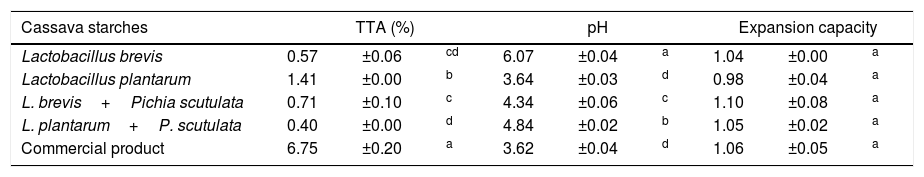
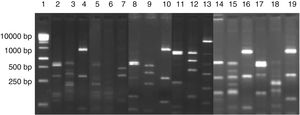
ResultsIdentification of microorganisms from cassava flour manufacturerLAB were present in all samples during a 56-day cassava fermentation with higher counts (5.8–7.9log10CFU/g) than those obtained for yeasts (1.7–7.8log10CFU/g), which were found from day 0 to 26. The 79 isolates of LAB identified were Gram-positive, catalase negative and mostly rods (83%). The bacterial isolates were efficiently grouped by RFLP (Figs. 1 and 2) in twelve different species. Seven species of Lactobacillus were isolated: L. brevis (n=17, 21.5%), L. fermentum (n=12, 15.2%), L. plantarum (n=11, 13.9%), Lactobacillus casei/Lactobacillus paracasei (n=7, 8.9%), Lactobacillus harbinensis (n=5, 6.3%), Lactobacillus parabuchneri (n=4, 5.0%) and Lactobacillus ghanensis (n=1, 1.3%). Enterococcus faecium (n=13, 16.5%) was also isolated and, in minor proportions, Weisella cibaria (n=3, 3.8%), Lactococcus garvieae (n=3, 3.8%), Lactococcus lactis subsp. lactis (n=2, 2.5%) and L. mesenteroides (n=1, 1.3%). The most frequent isolates were L. brevis, isolated from six out of seven sample collections (5.8–7.5log10CFU/g), and L. plantarum, isolated in five sample collections (5.4–6.4log10CFU/g).
Digestion profile of lactic acid bacteria isolated from cassava fermentation in a cassava flour manufacturer. Lanes: 1: 1kb DNA ladder; 2–4: Lactobacillus brevis profile (2-MspI, 3-HaeIII, 4-HinfI); 5–7: Lactobacillus fermentum profile (5-MspI, 6-HaeIII, 7-HinfI); 8–10: Lactobacillus plantarum profile (8-MspI, 9-HaeIII, 10-HinfI); 11–13: Lactobacillus casei/L. paracasei profile (11-MspI, 12-HaeIII, 13-HinfI); 14–16: Lactobacillus harbinensis (14-MspI, 15-HaeIII, 16-HinfI); 17–19: Lactobacillus parabuchneri profile (17-MspI, 18-HaeIII, 19-HinfI).
Digestion profile of lactic acid bacteria isolated from cassava fermentation in a cassava flour manufacturer. Lanes: 1: 1kb DNA ladder; 2–4: Lactobacillus ghanensis profile (2-MspI, 3-HaeIII, 4-HinfI); 5–7: Enterococcus faecium profile (5-MspI, 6-HaeIII, 7-HinfI); 8–10: Weisella cibaria profile (8-MspI, 9-HaeIII, 10-HinfI); 11–13: Lactococcus garvieae profile (11-MspI, 12-HaeIII, 13-HinfI);14–16: Lactobacillus lactis profile (14-MspI, 15-HaeIII, 16-HinfI); 17–19: Leuconostoc mesenteroides profile (17-MspI, 18-HaeIII, 19-HinfI).
All of the eleven isolates that were presumably identified by 16S rRNA sequence analysis as belonging to the L. plantarum group were posteriorly confirmed as species of L. plantarum by Multiplex PCR Assay using recA Gene-Derived Primers.
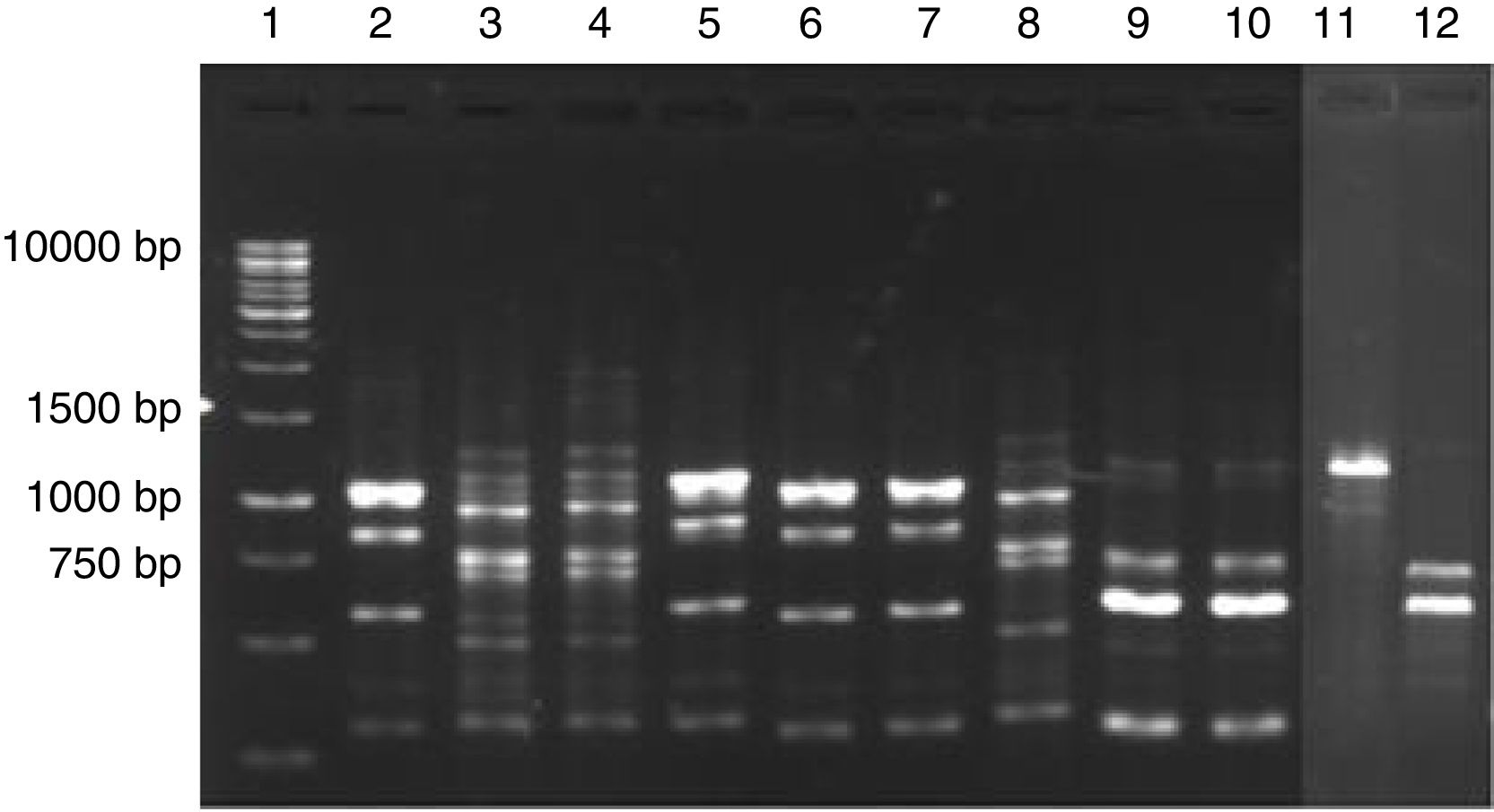
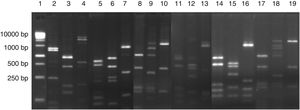
The 23 yeast isolates were efficiently grouped by analysis of their molecular profiles obtained by PCR fingerprinting (Fig. 3). The most commonly isolated yeast species were Pichia scutulata (n=12, 52.2%) and Kazachstania exigua (n=4, 17.4%), both found from day 0 to 12 (>4log10CFU/g). Candida humilis (n=3, 13.0%), Geotrichum fragrans (n=3, 13.0%) and Candida ethanolica (n=1, 4.4%) were also isolated.
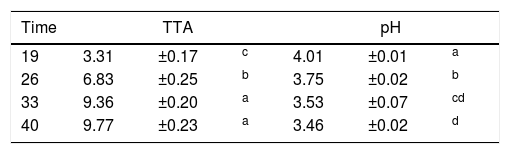
Evaluation of samples from cassava flour manufacturerThe pH values decreased and there was a consequent increase in TTA (Table 1). Bacillus spp. were found in the final product sample, with counts of 9.33×102CFU/g, which is a value lower than the tolerance permitted by Brazilian law (3.00×103CFU/g).31 Neither thermotolerant coliforms nor Salmonella spp. were detected.
TTA and pH of samples from a cassava flour manufacturer in the municipality of Formiga, Minas Gerais (MG) state, Brazil.
| Time | TTA | pH | ||||
|---|---|---|---|---|---|---|
| 19 | 3.31 | ±0.17 | c | 4.01 | ±0.01 | a |
| 26 | 6.83 | ±0.25 | b | 3.75 | ±0.02 | b |
| 33 | 9.36 | ±0.20 | a | 3.53 | ±0.07 | cd |
| 40 | 9.77 | ±0.23 | a | 3.46 | ±0.02 | d |
Mean values±standard deviation in the same column followed by different superscript letters are significantly different (p<0.001).
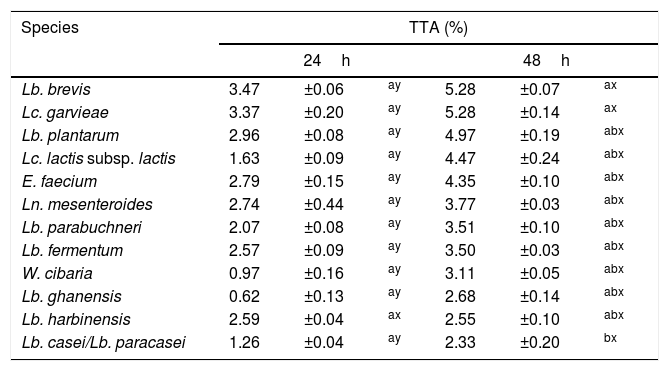
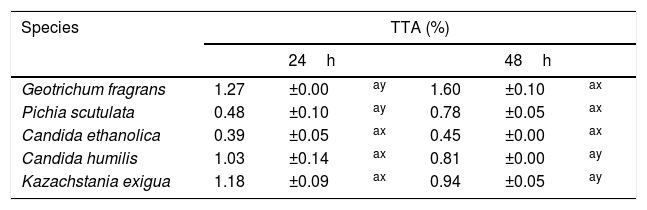
None of the 82 LAB strains isolated exhibited amylolytic activity. However, out of the 33 yeast strains isolated all the ones belonging to the species P. scutulata, C. humilis and C. ethanolica showed positive results (data not shown) for starch degradation (indicated by a clear zone of inhibition >1mm around colonies). TTA values evaluated for the predominant microbiota were greater at 48h than at 24h of fermentation for most of the LAB. The bacteria with higher acidity values were L. brevis and L. garvieae. Tables 2 and 3 show the results of TTA for the best strain isolated of each species.
TTA after 24 and 48h of fermentation with lactic acid bacteria isolated from a cassava flour manufacturer in the municipality of Formiga, Minas Gerais (MG) state, Brazil.
| Species | TTA (%) | |||||
|---|---|---|---|---|---|---|
| 24h | 48h | |||||
| Lb. brevis | 3.47 | ±0.06 | ay | 5.28 | ±0.07 | ax |
| Lc. garvieae | 3.37 | ±0.20 | ay | 5.28 | ±0.14 | ax |
| Lb. plantarum | 2.96 | ±0.08 | ay | 4.97 | ±0.19 | abx |
| Lc. lactis subsp. lactis | 1.63 | ±0.09 | ay | 4.47 | ±0.24 | abx |
| E. faecium | 2.79 | ±0.15 | ay | 4.35 | ±0.10 | abx |
| Ln. mesenteroides | 2.74 | ±0.44 | ay | 3.77 | ±0.03 | abx |
| Lb. parabuchneri | 2.07 | ±0.08 | ay | 3.51 | ±0.10 | abx |
| Lb. fermentum | 2.57 | ±0.09 | ay | 3.50 | ±0.03 | abx |
| W. cibaria | 0.97 | ±0.16 | ay | 3.11 | ±0.05 | abx |
| Lb. ghanensis | 0.62 | ±0.13 | ay | 2.68 | ±0.14 | abx |
| Lb. harbinensis | 2.59 | ±0.04 | ax | 2.55 | ±0.10 | abx |
| Lb. casei/Lb. paracasei | 1.26 | ±0.04 | ay | 2.33 | ±0.20 | bx |
Mean values±standard deviation in the same line followed by different superscript letters (x, y) are significantly different (p<0.001). Mean values ± standard deviation in the same column followed by different superscript letter (a, b) are significantly different (p<0.001).
TTA after 24 and 48h of fermentation with yeasts isolated from a cassava flour manufacturer in the municipality of Formiga, Minas Gerais (MG) state, Brazil.
| Species | TTA (%) | |||||
|---|---|---|---|---|---|---|
| 24h | 48h | |||||
| Geotrichum fragrans | 1.27 | ±0.00 | ay | 1.60 | ±0.10 | ax |
| Pichia scutulata | 0.48 | ±0.10 | ay | 0.78 | ±0.05 | ax |
| Candida ethanolica | 0.39 | ±0.05 | ax | 0.45 | ±0.00 | ax |
| Candida humilis | 1.03 | ±0.14 | ax | 0.81 | ±0.00 | ay |
| Kazachstania exigua | 1.18 | ±0.09 | ax | 0.94 | ±0.05 | ay |
Mean values±standard deviation in the same line followed by different superscript letters (x, y) are significantly different (p<0.001). Mean values±standard deviation in the same column followed by different superscript letter (a) are not significantly different (p>0.05).
One isolate of L. brevis and one isolate of L. plantarum were selected to be tested as the starter cultures because they were among the predominant LAB that had higher values of acidity. The isolate of yeast chosen to be tested as starter culture was one strain of P. scutulata, the predominant yeast species, which exhibited amylolytic activity. Moreover, P. scutulata ferments glucose, which is important for the production of secondary metabolites such as volatile compounds responsible for the characteristic aroma of sour cassava starch. In the present study, we tested four starter cultures: L. brevis; L. plantarum; L. brevis in a mixed culture with P. scutulata; and L. plantarum in a mixed culture with P. scutulata.
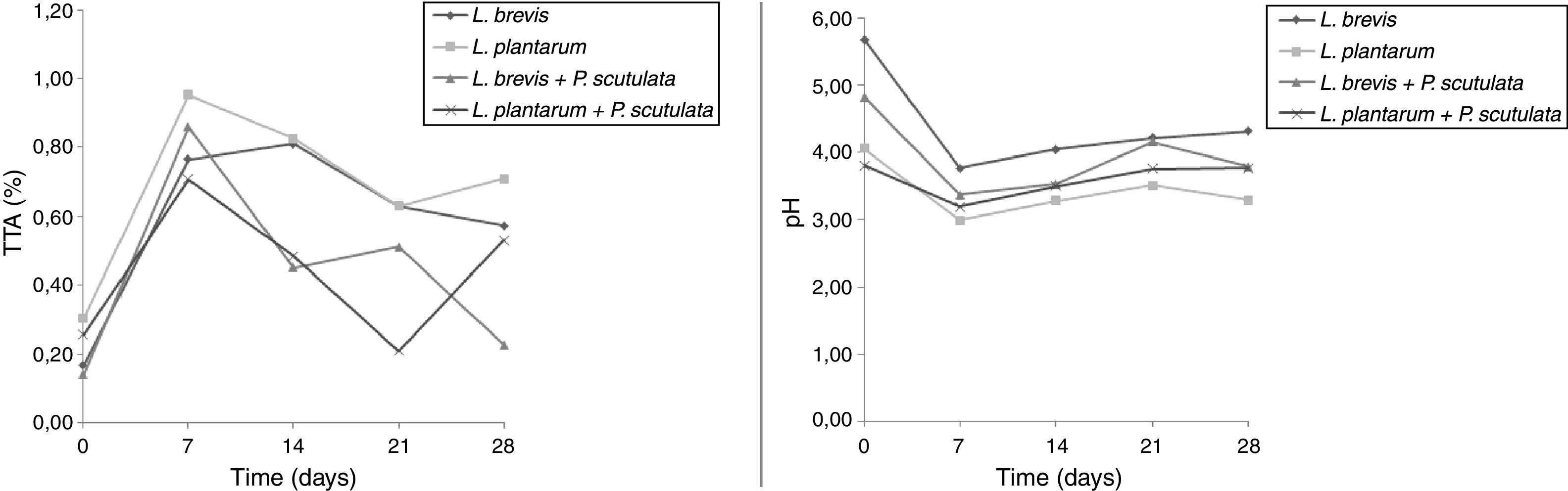
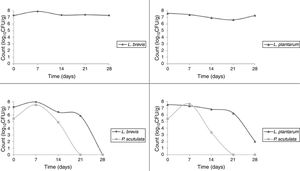
Use of starter cultures for sour cassava starch processingMonitoring of TTA and pH during the pilot-scale fermentation process revealed that values obtained for different starter cultures did not differ (Fig. 4). TTA ranged from 0.14% to 0.71%. The pH ranged from 5.69 to 3.29. ANOVA showed that the extent of fermentation significantly changed the TTA and pH of cassava starch.
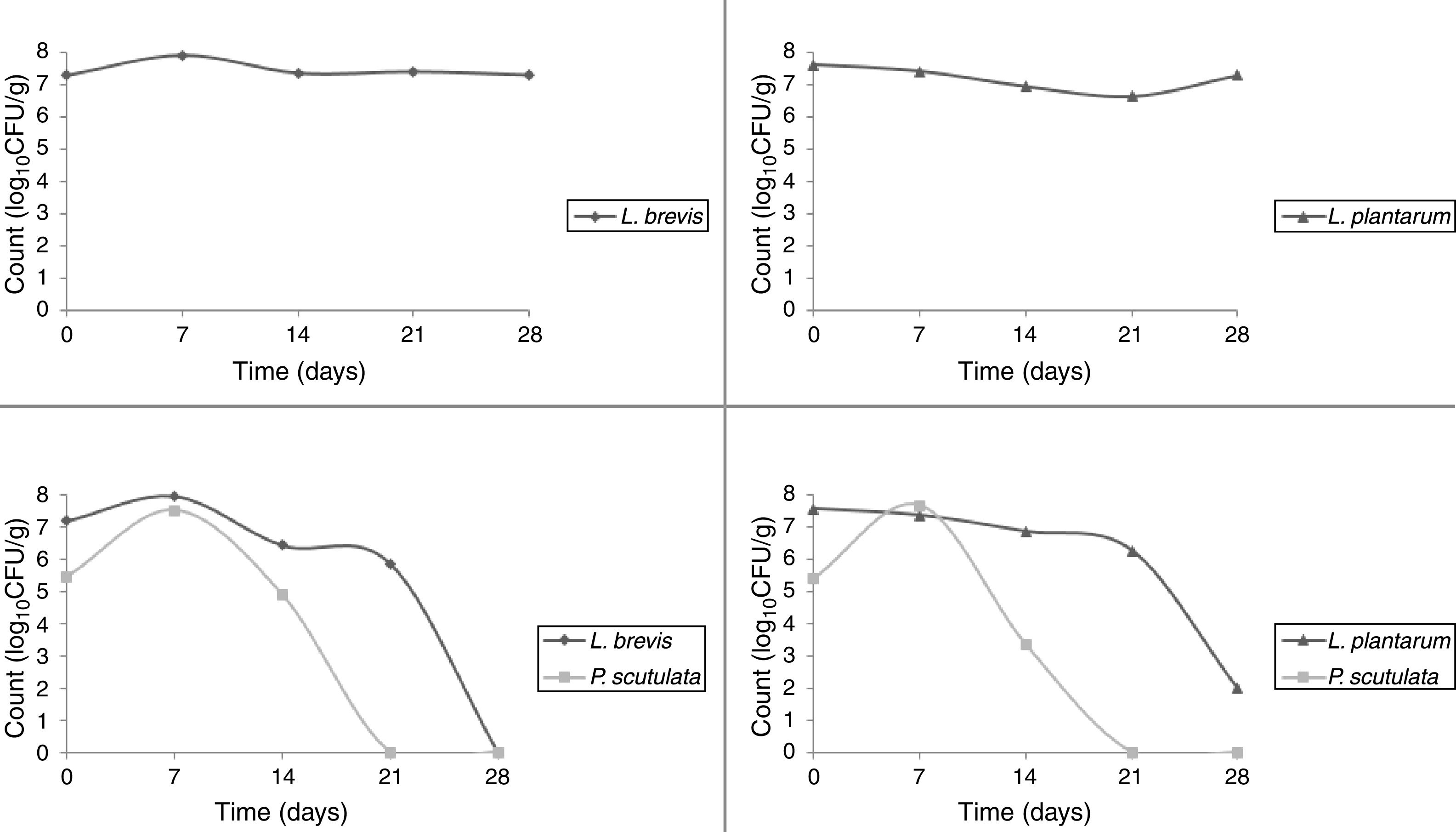
The starter cultures used were able to prevail during cassava pilot-scale fermentation; therefore, all the isolates selected for identification did correspond to the starter cultures inoculated. The LAB was observed to remain in fermentation during 28 days. It was observed that P. scutulata showed a maximum count, approximately 7log10CFU/g, within 7 days of fermentation, and the growth decreased reaching counts between 3 and 4log10CFU/g with 14 days. The number of viable cells of L. brevis was higher than the number of viable cells of L. plantarum when in association with the yeast. On the 21st day, the presence of the yeast was no longer observed (Fig. 5).
Cassava starch, the dried final product, obtained with L. plantarum exhibited the highest TTA value among the samples. Although this value was not close to the value of commercial product, it was within the established limits of legislation, which mandates a maximum acidity of 1.0% for sweet cassava starch and a maximum acidity of 5.0% for sour cassava starch.32 Hence, the product obtained with L. plantarum single culture could be called sour cassava starch. Final products obtained with other starter cultures might be called sweet cassava starch. The pH value of starch obtained with L. plantarum single culture was the only value that did not differ from the pH value of the commercial product. The expansion capacities for the cassava starches obtained with the starter cultures and for commercial product did not differ (Table 4). Cassava starches produced via a pilot-scale fermentation process proved to be within the microbiological limits31 and were therefore suitable for human consumption.
TTA, pH and expansion capacity for cassava starches (dried final products) obtained in a pilot scale fermentation compared with a commercial product.
| Cassava starches | TTA (%) | pH | Expansion capacity | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Lactobacillus brevis | 0.57 | ±0.06 | cd | 6.07 | ±0.04 | a | 1.04 | ±0.00 | a |
| Lactobacillus plantarum | 1.41 | ±0.00 | b | 3.64 | ±0.03 | d | 0.98 | ±0.04 | a |
| L. brevis+Pichia scutulata | 0.71 | ±0.10 | c | 4.34 | ±0.06 | c | 1.10 | ±0.08 | a |
| L. plantarum+P. scutulata | 0.40 | ±0.00 | d | 4.84 | ±0.02 | b | 1.05 | ±0.02 | a |
| Commercial product | 6.75 | ±0.20 | a | 3.62 | ±0.04 | d | 1.06 | ±0.05 | a |
Mean values±standard deviation in the same column followed by different superscript letters are significantly different (p<0.001).
During the isolation of LAB and yeasts from cassava fermentation, LAB were present in all samples collected with higher counts than those obtained for yeasts. Many authors also reported the presence of LAB as prevalent microorganisms in cassava fermentations.33–35 High LAB counts are evidence of the importance of these microorganisms in cassava fermentation.
Among the LAB often found in traditional cassava fermentations, many authors report the presence of L. brevis and L. plantarum,7,9,10,34,36,37 which were among the most frequent isolates in the present study. Lacerda et al.6 reported L. plantarum and L. fermentum as the predominant LAB during sour cassava starch fermentation isolated from two cassava flour manufacturers located in the municipality of Conceição dos Ouros (MG, Brazil). L. brevis was isolated in minor proportions. The counts of these bacteria were similar to those found in this study. The microbiota from cassava fermentation has different origins and may come from raw materials, utensils and equipment used in its production. Insects or handlers can also carry these microorganisms.
The most commonly isolated yeast species, P. scutulata, K. exigua, C. humilis, G. fragrans and C. ethanolica were also found in other cassava fermentation studies. Lacerda et al.6 collected samples from sour cassava starch fermentation and identified the following yeast species: Pichia sp., genus that was formerly known as Issatchenkia sp., was found in counts of 5log10CFU/g; C. humilis and C. ethanolica were also frequently isolated. Wilfrid Padanou10 found a prevalence of Saccharomyces cerevisiae (22%) and P. scutulata (20%) in the fermentation of lafun, an African product obtained from cassava submerged fermentation. The occurrence of these yeasts suggests that they could contribute to the sensorial quality. Several studies revealed the presence of the genus Candida in cassava fermentations to obtain different products.6,7,10,38,39 In the present study, one species of the genus Pichia and two species of the genus Candida were isolated, showing that species of these genera can grow in acidic conditions found in cassava fermentation tanks.
The acidic environment created by LAB favors the proliferation of yeasts in foods. Simultaneously, the growth of LAB is stimulated by the presence of yeasts, which provide growth factors such as vitamins and nitrogen compounds.40 The association of LAB and yeasts during fermentation has a significant impact on food quality parameters such as texture, flavor and nutritional values.41 Even though the complex interactions between LAB and yeasts are not yet fully understood, it is known that LAB and yeasts have the ability to adapt to many different substrates.42
The decrease of pH values and the consequent increase in TTA of samples from cassava flour manufacturer were expected results because, during fermentation, the synthesis of organic acids occurs, leading to acidification of the medium. It is possible that yeasts have a small contribution or do not contribute to acidification, especially because most yeasts are not resistant to extreme acidic conditions. The pH values found in the present study are lower than those determined by Coulin et al.7 in attiéké, a fermented cassava product. The average pH of this food during the traditional fermentation in small scale was 5.0.
According to the microbiological analyses, sour cassava starch from the fermentation tank studied would be considered suitable for human consumption. Verification of the presence of B. cereus is important because it produces toxins and may cause food poisoning when consumed in numbers higher than 105CFU/g. Besides the ability to produce organic acids, LAB also contribute to inhibition of pathogens due to production of antimicrobial compounds such as hydrogen peroxide, diacetyl and bacteriocins.43,44 LAB acidification power is known to be effective in controlling growth of microorganisms in food and extending its shelf life.45
The microorganisms considered as predominant in the present study were the ones with higher counts and the most frequently isolated during cassava fermentation process. The results are in agreement with the findings of predominant microorganisms in traditional cassava fermentations, including sour cassava starch fermentation, as reported in many previous studies.6–11,33–38,46–48 Therefore, the microorganisms isolated and identified in the present study are a suitable source of microorganisms for use in tests for the selection of starter cultures.
Selection of starter cultures showed that some yeast species exhibited amylolytic activity. This observation suggests that yeasts possibly play an important role at the beginning of fermentation, degrading starch and releasing sugars for the growth of LAB and their own. I also showed that TTA values obtained by LAB were higher than those obtained by yeasts, which indicates that the former are mainly responsible for acidification, which was also confirmed by Oguntoyinbo and Dodd.35
L. plantarum, selected as a starter culture for the production of sour cassava starch in pilot-scale fermentation process, has already been evaluated as a starter culture for the fermentation of other traditional cassava product. Kostinek et al.33 assessed the technological properties of the prevalent LAB in the fermentation of gari, an African product obtained by cassava solid-state fermentation, and noted that L. plantarum exhibited better and faster acid production among the isolated bacteria and was recommended as the starter culture. Huch et al.49 tested L. plantarum and concluded that its success to predominate in cassava fermentation demonstrates the potential for its development as a starter culture for gari industrialization. Edward et al.50 investigated the use of lyophilized LAB strains as starter cultures for gari production and found that L. plantarum could be produced at low cost.
Monitoring of TTA revealed values higher than the ones determined by Vogelmann et al.42 when studying cassava fermented by an association of LAB and yeasts used as starter cultures. The values found by the authors ranged from 10.3 to 11.5SH (Degrees Soxhlet-Henkel), which corresponds to 0.23% and 0.26%, respectively. Monitoring of pH revealed values similar to the ones determined by Vogelmann et al.,42 which varied from 4.1 to 4.2 during the 12 days of fermentation.
The monitoring of the viability of starter cultures showed that the cultures used were able to prevail during cassava pilot-scale fermentation. This result may suggest a strong association between LAB and yeasts at the beginning of fermentation and indicates that LAB play an important role during the entire process, especially concerning the quality and safety of the final product. Besides that, the use of starter cultures may optimize the fermentation process decreasing the fermentation time. Huch et al.49 tested L. plantarum and L. fermentum as starter cultures for the production of gari. The molecular monitoring by Random Amplified Polymorphic DNA (RAPD)-PCR and typing techniques by Pulsed Field Gel Electrophoresis (PFGE) indicated that L. plantarum was successful in asserting itself as a predominant strain, which did not happen with L. fermentum.
L. plantarum showed better performance as a starter culture for sour cassava starch production in a pilot-scale fermentation process. Cassava starch produced by this strain was found to have the highest value of TTA when compared with the other starter cultures; the pH and expansion capacity were not different from the values obtained by the commercial product; also, there was no evidence of pathogens. Therefore, further studies are required for the establishment of a starter culture that will contribute to the standardization of the cassava fermentation conditions, thereby ensuring higher quality products and consumer acceptability.
Conflicts of interestThe authors declare no conflicts of interest.