Introduction. Chronic hepatitis B (CHB) is still a public health problem and its mechanism remains unclear. In this study, we detect the skewness of T cell receptor beta chain variable gene (TCR Vβ) in peripheral blood lymphocytes (PBL) and the liver infiltrating lymphocytes (LIL) of patients with CHB; and hope to provide information for further research on the pathogenic mechanism of CHB.
Material and methods. Fifteen patients with CHB, ten healthy volunteers and three patients with liver cysts were recruited as the subjects. The usage of TCR Vβ of PBL and LIL were measured and compared; the associations of the TCR Vβ usage of PBL with some hematological indices, including human leukocyte antigen (HLA) alleles, percents of CD4+ and CD8+ T cells, sera levels of HBV-DNA and IFN-γ, were analyzed.
Results. In PBL, Vβ12 and Vβ13.1 were the highest predominant usage genes which usage frequencies were all 46.7%; Vβ23 was the key limited usage gene (40.0%). In LIL, the mainly predominant and limited usage gene was Vβ13.1 (73.3%) and Vβ23 (46.7%), respectively. About half of the patients with CHB with HLA-DR9 or HLA-DR12 showed the predominant usage of Vβ5.2 or Vβ13.2. In patients with CHB, the percentage of CD4+ T cells was 33.41 ± 5.39 %, that of CD8+ T cells was 28.67 ± 6.77 %; the concentration of IFN-γ was 182.52 ± 44.16 pg/mL. Compared to the healthy controls, there were significant differences for these data (P < 0.05). Neither ALT nor HBV-DNA was relative to the usage of TCR Vβ.
Conclusions. PBL and LIL share the common sknewness of TCR Vβ genes which probably relates to some hematological indices. However, the roles of such similarities and associations in the development of CHB need further study.
Chronic hepatitis B (CHB) - defined as persistence of hepatitis B surface antigen (HBsAg) for six months or more - is a major public health problem. Worldwide, there are an estimated 240 million chronically infected persons worldwide, and it is estimated that about 650,000 people will die annually due to CHB.1 Universal hepatitis B immunization programmes that target infants have been highly effective in reducing the incidence and prevalence of hepatitis B (HB) in many endemic countries. However, these programmes will not have an impact on HBV-related deaths until several decades after their introduction.1
Therefore, it is still a long-time and arduous task for the researchers to prevent and therapy HB. Sadly, the exact mechanisms of the disease are yet unclear today; fortunately, a viewpoint has been determined that the occurrence of liver injury is not caused by HBV itself but the cellular immune response.2 Among the process of immune response, T cell plays the most important role.3,4 As to the mechanism, several scholars considered that some cytokines secreted by HBV-activated T cells in the process of clearing HBV probably brought damage to liver cells.5 Presently, the relationship between the changes of T cell receptor (TCR) and HBV infection became a hot research topic.
As we known, T cells recognize the complex of antigens peptide and human leukocyte antigen (HLA) through TCR, which is composed mainly of receptor α and β chains (more than 95%). The third complementarity determining region (CDR3) has been defined for the variable regions of beta chains (Vβ). Functionally, Vβ genes associate with antigen-recognition process, and the part of the TCR Vβ mainly responsible for the specific interaction with the antigenic peptide is CDR3.6,7 T cells of different specificity express different CDR3 which vary in length or sequence.8,9 The specific recognition to antigen peptides might result in clonal expansion of the T cells, and in which TCR Vβ CDR3 exhibit special changes. Therefore, measuring the frequency of specific CDR3 sequences could reflect the degree of T cell clone. Accordingly, analysis of CDR3 size distribution has been used to define the degree of clonality of T cells in response to the special antigens.10 To date, the studies of the skewness of TCR Vβ genes have involved some diseases, such as type 1 diabetes,11 colorectal carcinoma,12 tuberculosis,13 and so on. The usage of TCR Vβ genes relates to HB has also been reported. Wu SQ, et al.14 reported that Vβ13.1, Vβ17 and Vβ22 were restrictedly used in some CHB patients, and suggested that the three predominant genes probably were the special clones to HB. In the study of Shi WJ, et al., 15 the expression levels of Vβ1, Vβ12 and Vβ20 of the patients with fulminant hepatitis B (FHB) were significantly higher than those of healthy controls, while the levels of Vβ5, Vβ7, Vβ13, Vβ14, Vβ15, Vβ22 and Vβ23 of the patients were lower than those of the controls. Accordingly, the authors thought that these Vβ genes probably related to the pathogenesis of the liver inflammation process of FHB. The two reports were different form another reference, in which Vβ8, Vβ11, Vβ13, Vβ20 and Vβ24 were frequently used in the patients with chronic asymptomatic hepatitis B virus infection.16 Although the above studies exhibit some significant findings, there are several limitations among them. For example, the skewed TCR Vβ genes are always different in different reports, and then, which is the real factor for the pathogenesis of HB still remains unclear; all the studies generally focus on the skewness of TCR V genes of the peripheral blood, how about those in liver tissue is rarely revolved. The existence of such limitations indicates that there will lots of work relate to TCR Vβ gene usage to do in future.
In the previous reports,6,7,17 with real-time florescence quantitative polymerase chain reaction (RQ-PCR) and melting curve analysis technique (MCAT), we successfully detected the skewness of several diseases. In this study, we would assay the TCR Vβ gene usage of peripheral blood lymphocyte (PBL) and liver infiltrating lymphocyte (LIL) of patients with CHB with the same method, and hope to provide information for the research on the pathogenesis of CHB through comparatively analyzing the clone features of the two specimens.
Material and MethodsPatientsFifteen patients with CHB (as shown in Table 1) were diagnosed according to the Guideline on Prevention and Treatment of CHB (2010 version).18 Ten healthy volunteers and three patients with liver cysts were recruited as controls who provided peripheral blood and liver tissue samples, respectively. All subjects had not been treated with immunomodulating drugs in the six months prior to the study and were seronegative for markers of the other hepatitis viruses (including hepatitis A, C, D and E virus), HIV and other pathogenic infections. Excluded from the study were patients with tumors and immunological disorders. Written informed consents were obtained from all the participants. This study protocol was approved by the Hospital Ethics Committee.
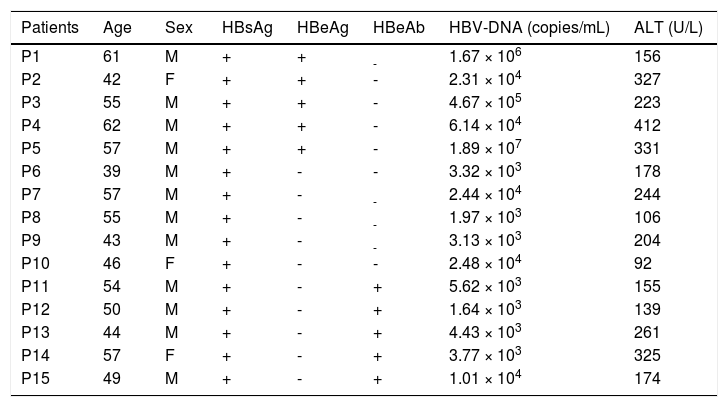
The basic clinical information of all the patients with chronic hepatitis B.
| Patients | Age | Sex | HBsAg | HBeAg | HBeAb | HBV-DNA (copies/mL) | ALT (U/L) |
|---|---|---|---|---|---|---|---|
| P1 | 61 | M | + | + | - | 1.67 × 106 | 156 |
| P2 | 42 | F | + | + | - | 2.31 × 104 | 327 |
| P3 | 55 | M | + | + | - | 4.67 × 105 | 223 |
| P4 | 62 | M | + | + | - | 6.14 × 104 | 412 |
| P5 | 57 | M | + | + | - | 1.89 × 107 | 331 |
| P6 | 39 | M | + | - | - | 3.32 × 103 | 178 |
| P7 | 57 | M | + | - | - | 2.44 × 104 | 244 |
| P8 | 55 | M | + | - | - | 1.97 × 103 | 106 |
| P9 | 43 | M | + | - | - | 3.13 × 103 | 204 |
| P10 | 46 | F | + | - | - | 2.48 × 104 | 92 |
| P11 | 54 | M | + | - | + | 5.62 × 103 | 155 |
| P12 | 50 | M | + | - | + | 1.64 × 103 | 139 |
| P13 | 44 | M | + | - | + | 4.43 × 103 | 261 |
| P14 | 57 | F | + | - | + | 3.77 × 103 | 325 |
| P15 | 49 | M | + | - | + | 1.01 × 104 | 174 |
M: male. F: female. +: positive. -: negative,
PBL and LIL were isolated from peripheral blood and liver tissue samples by Ficoll-Hypaque density centrifugation, respectively. Total RNA was extracted using an Omega RNA extraction kit according to the manufacturer’s instructions. 3 /µg total RNA was reverse transcribed with 250 pm olig (dT), 200 U Moloney murine leukemia virus reverse transcriptase, and 5 /xL of 10 mM dNTP mix (cDNA Synthesis Kit; MBI-Fermentas) in a eppendorf tube. The total volume was 50 /xL. Six reactions were performed for each sample.
Detection of TCR Vβ usageThe sense primer and anti-sense primer for 24 TCR Vβ genes families (both of Vβ5 and Vβ13 include two subfamilies: Vβ5.1 and Vβ 5.2, Vβ13.1 and Vβ13.2) were previously described.6,7,17 With RQ-PCR, 24 TCR Vβ genes usage was detected, and the detail procedure was as following: 2 µL sense primer and anti-sense primer, 2 µL MgCl2 (2.0 µM), 5 µL dNTP (10 mM), 5 µL 10x buffer, 2 µL cDNA template, and 1.4 U Taq-polymerase were mixed, followed by PCR under the conditions: 94°C for 5 min, 94°C melting for 1 min, primer annealing at 56°C for 1 min, and 72°C for 3 min, 35 cycles; then extension at 72°C for 12 min. Finally, PCR products of 24 TCR Vβ genes were analyzed with MCAT.
Hla Analysis5 mL of blood was taken from each of the subjects, and with which HLA-A and HLA-DR were detected by polymerase chain reaction with sequences-specific primers (PCR-SSP). The reagents were all bought from Shanghai Shenggong Bioengineer Ltd., China.
Detection of homological indicesThe percents of CD4+ and CD8+ T cells were detected with flow cytometer (BD FACSCaliber, USA). HBV-DNA was assayed with PCR instrument (ABI1500, USA), and the concentration of IFN-γ was detected with ELISA kit (Sigma, Singapore). All the experiments were performed strictly according to the manufacturers instructions.
Statistical analysisTCR Vβ usage of PBL was compared with that of LIL. The comparison was performed according to two calculation formulas.1
- •
Usage frequency (%) = N1/N0 × 100%. N1 meant the total number of certain a TCR Vβ gene family which exhibited advantage and/or limited usage; N0 represented the total number of the same TCR Vβ genes.
- •
Coincidence rate (%) = 2A/B × 100%.
A represented the total number of TCR Vβ genes which shared advantage (limited) usage in PBL and LIL; B represented the total number of all biased TCR Vβ genes in the two specimens. All the data were analyzed with the SPSS statistic software (version 15.0). %2 test was used to determine the difference of coincidence rates or the frequencies of HLA alleles. The level of 0.05 was taken as the criteria for significance.
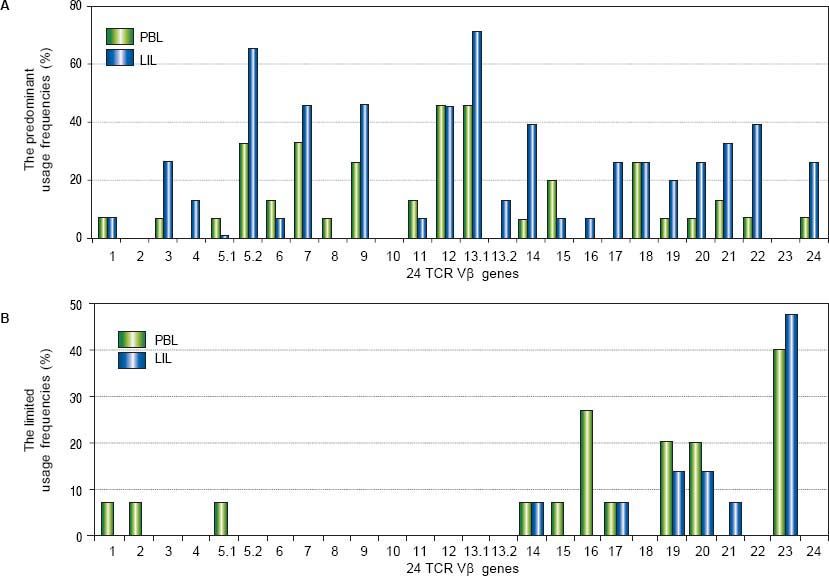
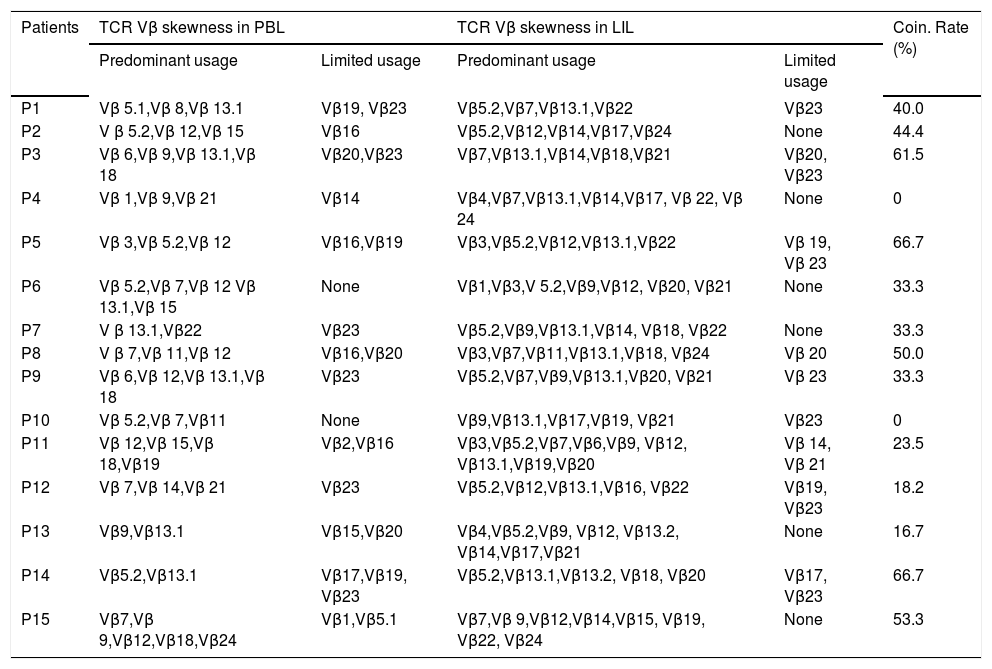
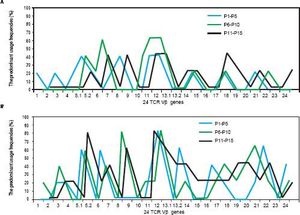
ResultsTCR Vβ usages of PBLOf all the patients with CHB, some TCR Vβ genes showed predominant and limited usage. The genes of highest predominant usage were Vβ12 and Vβ13.1, and the usage frequencies all were 46.7%. Vβ5.2 and Vβ7 were next to it with the same frequency of 33.3%. The most limited usage gene was Vβ23 (40.0%) which followed by Vβ16 (26.7%) (Table 2, Figure 1). In the ten healthy volunteers and three patients with liver cyst, there was no skewed TCR Vβ gene.
The skewed TCR Vβ genes in PBL and LIL of the patients with CHB.
| Patients | TCR Vβ skewness in PBL | TCR Vβ skewness in LIL | Coin. Rate (%) | ||
|---|---|---|---|---|---|
| Predominant usage | Limited usage | Predominant usage | Limited usage | ||
| P1 | Vβ 5.1,Vβ 8,Vβ 13.1 | Vβ19, Vβ23 | Vβ5.2,Vβ7,Vβ13.1,Vβ22 | Vβ23 | 40.0 |
| P2 | V β 5.2,Vβ 12,Vβ 15 | Vβ16 | Vβ5.2,Vβ12,Vβ14,Vβ17,Vβ24 | None | 44.4 |
| P3 | Vβ 6,Vβ 9,Vβ 13.1,Vβ 18 | Vβ20,Vβ23 | Vβ7,Vβ13.1,Vβ14,Vβ18,Vβ21 | Vβ20, Vβ23 | 61.5 |
| P4 | Vβ 1,Vβ 9,Vβ 21 | Vβ14 | Vβ4,Vβ7,Vβ13.1,Vβ14,Vβ17, Vβ 22, Vβ 24 | None | 0 |
| P5 | Vβ 3,Vβ 5.2,Vβ 12 | Vβ16,Vβ19 | Vβ3,Vβ5.2,Vβ12,Vβ13.1,Vβ22 | Vβ 19, Vβ 23 | 66.7 |
| P6 | Vβ 5.2,Vβ 7,Vβ 12 Vβ 13.1,Vβ 15 | None | Vβ1,Vβ3,V 5.2,Vβ9,Vβ12, Vβ20, Vβ21 | None | 33.3 |
| P7 | V β 13.1,Vβ22 | Vβ23 | Vβ5.2,Vβ9,Vβ13.1,Vβ14, Vβ18, Vβ22 | None | 33.3 |
| P8 | V β 7,Vβ 11,Vβ 12 | Vβ16,Vβ20 | Vβ3,Vβ7,Vβ11,Vβ13.1,Vβ18, Vβ24 | Vβ 20 | 50.0 |
| P9 | Vβ 6,Vβ 12,Vβ 13.1,Vβ 18 | Vβ23 | Vβ5.2,Vβ7,Vβ9,Vβ13.1,Vβ20, Vβ21 | Vβ 23 | 33.3 |
| P10 | Vβ 5.2,Vβ 7,Vβ11 | None | Vβ9,Vβ13.1,Vβ17,Vβ19, Vβ21 | Vβ23 | 0 |
| P11 | Vβ 12,Vβ 15,Vβ 18,Vβ19 | Vβ2,Vβ16 | Vβ3,Vβ5.2,Vβ7,Vβ6,Vβ9, Vβ12, Vβ13.1,Vβ19,Vβ20 | Vβ 14, Vβ 21 | 23.5 |
| P12 | Vβ 7,Vβ 14,Vβ 21 | Vβ23 | Vβ5.2,Vβ12,Vβ13.1,Vβ16, Vβ22 | Vβ19, Vβ23 | 18.2 |
| P13 | Vβ9,Vβ13.1 | Vβ15,Vβ20 | Vβ4,Vβ5.2,Vβ9, Vβ12, Vβ13.2, Vβ14,Vβ17,Vβ21 | None | 16.7 |
| P14 | Vβ5.2,Vβ13.1 | Vβ17,Vβ19, Vβ23 | Vβ5.2,Vβ13.1,Vβ13.2, Vβ18, Vβ20 | Vβ17, Vβ23 | 66.7 |
| P15 | Vβ7,Vβ 9,Vβ12,Vβ18,Vβ24 | Vβ1,Vβ5.1 | Vβ7,Vβ 9,Vβ12,Vβ14,Vβ15, Vβ19, Vβ22, Vβ24 | None | 53.3 |
PBL: peripheral blood lymphocyte. LIL: liver infiltrating lymphocyte. UF: usage frequency. CR: coincidence rate.
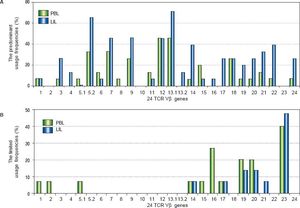
The comparison of the usage frequencies of 24 TCR Vβ genes of PBL with those of LIL in the patients with CHB. A. The comparison of the predominant usage frequencies of 24 TCR Vβ genes of PBL with those of LIL. B. The comparison of the limited usage frequencies of 24 TCR Vβ genes of PBL with those of LIL.
In the patients with CHB, there were some TCR Vβ genes showed skewed. Vβ 13.1 was the highest predominant usage gene (73.3%) which followed by Vβ 5.2 (66.7%). Vβ 23 was the limited usage gene with the highest frequency (46.7%); Vβ19 and Vβ 20 were next to it and the both frequencies were 20.0% (Table 2, Figure 1). In the three patients with liver cysts, no skewed TCR Vβ gene was found.
Comparison of TCR Vβ usages of PBL and LILThe coincidence rates between the skewness of TCR Vβ genes of PBL and that of LIL ranged from 0% to 66.7% (Table 2). The similarities shared in the two samples included three aspects:
- •
Vß12 and Vß 13.1 were the highest predominant usage genes.
- •
Vß 10 and Vß 23 genes were never predominantly used.
- •
Vß 19, Vß 20 and Vß 23 were the common limited usage genes.
The differences also contained three points:
- •
The number of preferential genes in LIL was larger than that in PBL.
- •
The predominant usage frequencies of many TCR Vβ genes of LIL were significantly higher than those of PBL.
- •
The number of the restricted usage genes of PBL was more than that of LIL.
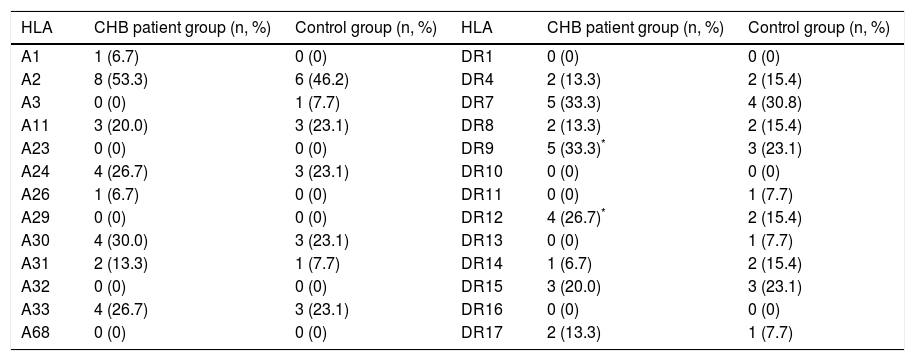
As shown in table 3, HLA-A2 was the gene with the highest frequency (53.3%) in the patients with CHB; the corresponding data in the control group was 46.2%.
The frequencies of HLA-A and HLA-DR in CHB patient and control groups.
| HLA | CHB patient group (n, %) | Control group (n, %) | HLA | CHB patient group (n, %) | Control group (n, %) |
|---|---|---|---|---|---|
| A1 | 1 (6.7) | 0 (0) | DR1 | 0 (0) | 0 (0) |
| A2 | 8 (53.3) | 6 (46.2) | DR4 | 2 (13.3) | 2 (15.4) |
| A3 | 0 (0) | 1 (7.7) | DR7 | 5 (33.3) | 4 (30.8) |
| A11 | 3 (20.0) | 3 (23.1) | DR8 | 2 (13.3) | 2 (15.4) |
| A23 | 0 (0) | 0 (0) | DR9 | 5 (33.3)* | 3 (23.1) |
| A24 | 4 (26.7) | 3 (23.1) | DR10 | 0 (0) | 0 (0) |
| A26 | 1 (6.7) | 0 (0) | DR11 | 0 (0) | 1 (7.7) |
| A29 | 0 (0) | 0 (0) | DR12 | 4 (26.7)* | 2 (15.4) |
| A30 | 4 (30.0) | 3 (23.1) | DR13 | 0 (0) | 1 (7.7) |
| A31 | 2 (13.3) | 1 (7.7) | DR14 | 1 (6.7) | 2 (15.4) |
| A32 | 0 (0) | 0 (0) | DR15 | 3 (20.0) | 3 (23.1) |
| A33 | 4 (26.7) | 3 (23.1) | DR16 | 0 (0) | 0 (0) |
| A68 | 0 (0) | 0 (0) | DR17 | 2 (13.3) | 1 (7.7) |
In comparison, the difference was not significant (P > 0.05). The frequencies of HLA-DR9 and HLA-DR12 were 33.3% and 26.7% in the patients with CHB, respectively; both were significantly higher than those of the control group (P < 0.05). Combining these data with the results of the TCR Vβ usage, it is easy to find that more than half of cases which high expressed HLA-DR9 showed the predominant usage of TCR Vβ5.2, such as P2, P6 and P14; almost all the cases of HLA-DR12 positive predominantly expressed TCR Vβ13.1, e.g. P1, P6, P9 and P14.
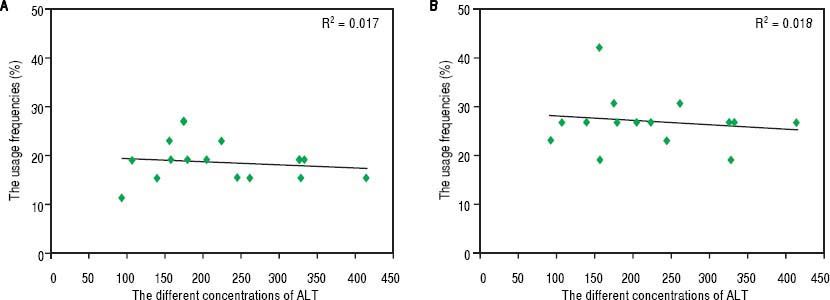
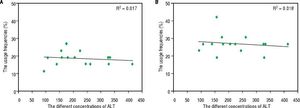
Hematological indices and TCR Vβ usageThe percentage of CD4+ T cells was 33.41 ± 5.39 % in the patients with CHB, and that was 38.9 ± 6.17 % in the healthy volunteers. Comparatively, there was significant difference between the rates of the two groups of subjects (P < 0.05). The ratio of CD8+ T cells was 28.67 ± 6.77 % and 23.88 ± 5.94% in patients with CHB and healthy volunteers, respectively; and there was a significant difference between the two rates (P < 0.05). The average concentration of serum IFN-y was 182.52 ± 44.16 pg/mL in patients with CHB, which was obviously higher than that of the healthy controls (14.87 ± 9.95 pg/mL, P < 0.001). Not only in PBL but also in LIL, there was no correlation between ALT and the skewness of TCR Vβ genes (Figure 3).
The relation between the serum levels of ALT and the usage frequencies of TCR Vβ of PBL (LIL) in patients with CHB. A. The relation between the serum levels of ALT and the usage frequencies of TCR Vβ of PBL. B. The relation between the serum levels of ALT and the usage frequencies of TCR Vβ of LIL.
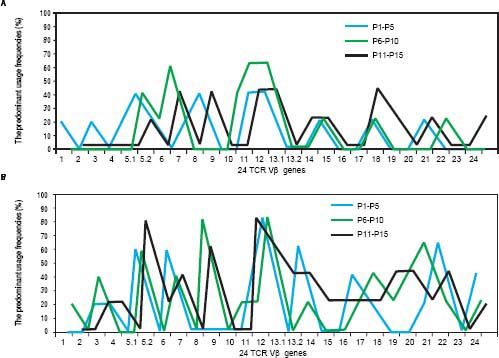
As shown in table 1, according to the detection results of HBsAg, HBeAg and HBeAb, HBV infection was divided into three serological patterns: HBsAg (+) & HBeAg (+) & HBeAb (−), HBsAg (+) & HBeAg (−) & HBeAb (−) and HBsAg (+) & HBeAg (−) & HBeAb (+). Not only in PBL but also in LIL, the predominant usage frequencies of TCR Vβ genes between the three patterns were similar (Figure 2). Besides, according to the comprehensive analysis of TCR Vβ gene usage and HBV-DNA copies, there was no positive association between the skewed TCR Vβ genes and HBV-DNA load.
DiscussionThe predominant usage of TCR Vβ in PBL from the patients with CHB has been ever described in a few reports. In Yao s study,19 Vβ8, Vβ12 and Vβ24 were the predominantly used genes. In another report, Vβ11 and Vβ12 highly expressed, and accordingly, the authors though that both genes probably associated with the occurrence of hepatitis B.20 In the present study, Vβ12 and Vβ13.1 were found as the most predominant usage genes in PBL. Obviously, Vβ12 was the common predominant usage gene in the three studies. This probably indicated tha,t Vβ12 was the just predominant gene with high specificity to CHB. As to Vβ13.1, Wu, et al.14 also found that it was frequently used in PBMC, and the usage frequency was high to 72.7% which was higher than the data of this study (46.7%). In another report, Vβ13 was also reported as the predominant usage gene,15 but because the researchers didn t identify it as Vβ 13.1 or Vβ 13.2, so we could not determine whether their results were consistent with ours. Compared to PBL, fewer studies focused on the dominant usage of TCR Vβ of LIL. In this study, most of Vβ genes showed predominant usage including Vβ 13.1 and Vβ 5.2. This was similar to Zhang s study,21 in which Vβ 13.1 was reported as the predominant usage gene with the highest frequency; Vβ 5.2 and Vβ 12 were next to it.
As to the limited usage Vβ genes, not only in PBL but also in LIL Vβ23 was always the highest frequent gene in this study. Dramatically, Vβ23 was the predominant usage gene in other several reports.14,16 According to our knowledge, such a difference probably reflected the individualization feature of the limited usage of TCR Vβ genes in patients with CHB.
To combine and analyze the skewness of TCR Vβ genes of PBL with that of LIL, it was easy to found that there were similarities and differences between the two specimens. In our opinions, the mechanisms for the similarities and differences probably focused on two aspects:
- •
Liver was the main place at where HBV located and proliferated, so the HBV antigen peptides in liver were much more than in peripheral blood. Naturally, as a key place for the cellular immune, there were more TCR Vβ genes skewed. This was the possible reason for what the total number of the skewed TCR Vβ genes in LIL was higher than that in PBL.
- •
Some T cells containing the skewed TCR Vβ genes of peripheral blood probably migrated from liver,22 so PBL and LIL shared the common skewed TCR Vβ genes. However, these reasoning needed further verifi-cation with more and powerful immunological or pathological experiments in future.
Recently, the association between HLA genes and the chronicity of HBV infection had been reported;23,24 but few studies focused on the relativity between HLA alleles and skewness of TCR Vβ genes of patients with CHB. In the present study, HLA alleles (HLA-A and HLA-DR) and TCR Vβ usage of the patients with CHB were simultaneously detected. In the results, there was no significant difference between the expression frequencies of HLA-A alleles of the patients with CHB and those of control group; while the frequencies of HLA-DR9 and HLA-DR12 of the CHB group were significantly higher than the data of the control group. This indicated that HLA-A gene probably played a small part in the chronicity of HBV infection; while HLA-DR was more important in such a progress. Besides, there was an interestingly phenomenon that the cases of HLA-DR9 or HLA-DR12 positive always showed the predominant usage of Vβ 5.2 or Vβ 13.1. This further strongly suggested that HLA-DR9 or HLA-DR12 were very likely to associate with the predominant usage of TCR Vβ genes. This finding was different from Sing’s reports22, in which HLA-B13, Bw4 and Cw3 associated with the oligoclonal use of Vβ 5.2, Vβ 11 and Vβ 17 gene families. However, both of studies showed that some HLA genes probably associated with the skewness of TCR Vβ genes for patients with CHB.
In the results of hematological indices, the percentage of CD4+ T cells of the patients was obviously lower than that of the healthy volunteers; while the rate of CD8+ T cells of the patients was significantly higher than the con-vtrols. Companying these results with the data relate to the skewness of TCR Vβ genes, we considered that the imbalance of T cell subpopulation probably was one of the reasons for the skewness of TCR Vβ genes. This was consistent with other scholars reports that TCR Vβ genes of the CD8+ T cells of patients with HBV infection exhibited a greater number of biased clones than CD4+ T cells.25,26 Besides, the average IFN-y concentration of the patients with CHB was much higher than that of healthy volunteers. This finding indicated that the level of IFN-y probably associated with the skewness of TCR Vβ genes of patients with CHB.27,28 But the high IFN-y concentration was the reason or the result for the skewness of TCR Vβ genes needed further study.
As shown in figure 2, the predominant usages of TCR Vβ of the patients with CHB were similar in the three serological patterns of infection. This probably because that the skewness of TCR Vβ genes kept a rather stable status under the long term of stimulation by HBV antigens; and moreover, such an antigen stimulation was uncorrelated with HBV-DNA load which can be known from the relationship between HBV-DNA copies and TCR Vβ gene usage. Besides, no relation could be found between ALT and TCR Vβ gene usage in the study. This was consistent with another reports,29 and both studies gave a suggestion that there was no obvious correlation between the TCR Vβ skewness and liver injury.
The comparison of the predominant usage frequencies of 24 TCR Vβ genes of PBL (LIL) in the patients with CHB between the three serological patterns of infection, which including HBsAg (+) & HBeAg (+) & HBeAb (−), HBsAg (+) & HBeAg (−) & HBeAb (−) and HBsAg (+) & HBeAg (−) & HBeAb (+). A. The comparison of the predominant usage frequencies of 24 TCR Vβ genes of PBL in the patients with CHB between the three serological patterns of infection. B. The comparison of the predominant usage frequencies of 24 TCR Vβ genes of LIL in the patients with CHB between the three serological patterns of infection.
In conclusion, this study showed two important findings. One was that there were common and different features for TCR Vβ usage in PBL and LIL of patients with CHB; the other was that some hematological indices probably associated with the skewness of TCR Vβ genes, including HLA-DR9, HLA-DR12, CD8+ T cell and IFNy. However, due to the small size of cases, there were still some questions need to be cleared up in future, such as the sequences of the skewed TCR Vβ genes, the relationship between TCR Vβ genes and each of HLA alleles, the mechanism for the effects of imbalance of CD4+ and CD8+ T on TCR Vβ gene usage, and so on.
AcknowledgementsAll the authors thank Dr. Xinsheng Yao for the guidance to the study.
Grant SupportThe work is granted by the Provincial Science and Technology Development Project (2012YD18054), the Provincial Nature Science Foundation (ZR2012HL29), the High School Science and Technology Plan Project (J11LF18), the Population and Family Planning Commission ([2011]13), and the Development Plan Project of Jining Science and Technology Bureau of Shandong Province ([2011] 57, 2014jnjc12 and 2014jnyx14 ).
Abbreviations- •
CDR3: the third complementarity-determining region.
- •
CHB: chronic hepatitis B.
- •
FHB: fulminant hepatitis B.
- •
HB: hepatitis B.
- •
HBsAg: hepatitis B surface antigen.
- •
HLA: human leukocyte antigen.
- •
LIL: liver infiltrating lymphocytes.
- •
MCAT: melting curve analysis technique.
- •
PBL: peripheral blood lymphocytes.
- •
RQ-PCR: real-time florescence quantitative polymerase chain reaction.
- •
TCR: T cell receptor.
- •
Vβ: beta chain variable gene.