Background and aim. The treatment efficacy of peginterferon plus ribavirin for patients with HCV genotype 1 is inferior to that in patients with HCV genotype 2, but the efficacy among patients with mixed HCV genotype 1 + 2 is less clear. We compared the treatment outcome of peginterferon alpha-2b plus ribavirin among naïve chronic hepatitis C patients in Taiwan with HCV genotype 1 and 2, and mixed genotype 1 + 2.
Material and methods. In this retrospective cohort study, 150 patients were treated with peginterferon alpha-2b once weekly, plus ribavirin, for 24 weeks. The endpoint was sustained virological response after receiving at least one dose of the study medication.
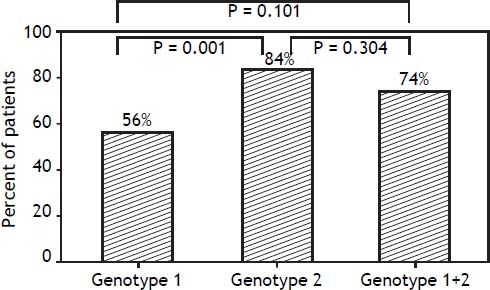
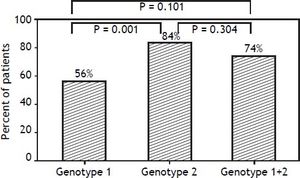
Results. There were no differences in clinical characteristics among the 3 groups. There were significant differences in rapid virological response rate between patients with genotype 1 and genotype 2 (64.7 vs. 85.5%, respectively; p < 0.05) and a sustained virological response rate (55.9 vs. 83.6%, respectively; p = 0.001). The rapid virological response rate differed between the genotype 1 and mixed genotype 1 + 2 groups (64.7 vs. 85.2%, respectively; p < 0.05), but the sustained virological response rate was similar (55.9 vs. 74.1%; p = 0.101). Conclusions. Using peginterferon alpha-2b plus ribavirin for 24 weeks to treat patients with HCV genotype 1 + 2 achieved a 74.1% sustained virological response rate; the treatment efficacy was not inferior to patients with HCV genotype 1, but the percentage of liver cirrhosis in mixed genotype 1 + 2 group was higher to 22%, it is worth to be appropriately valued and studied.
Despite widespread treatment, hepatitis C deaths are increasing, mostly due to inadequate detection and treatment.1 This is true even in developed countries. Accurate hepatitis C virus (HCV) geno-typing is very important for determining optimal strategies and for predicting or estimating the response rate after antiviral treatment.2
HCV can be classified into six major genotypes and numerous subtypes.3 The distribution of HCV genotypes varies by geographical location and groups at risk for infection in areas where genotype 1 and genotype 2 are distributed globally, including Taiwan.4
In some special groups, hepatitis C co-infection with human immunodeficiency virus (HIV) or hepatitis B virus (HBV) is an important issue. While the treatment policy is well known, less is known about treating the individual with mixed infection, including those with two or more HCV genotypes. In some HCV-infected patients, more than one genotype can be detected (mixed genotype infection).5 Recently, an HCV subtyping assay was developed; this assay uses a reliable polymerase chain reaction method to identify individual mixed genotype HCV infection.6,7 The global reported incidence of mixed genotype infection in chronic hepatitis C varies from 5% to 22%.8–10
We conducted a retrospective cohort study in our clinical practice to evaluate the role of HCV mixed genotype 1 + 2 on the treatment response to 24 weeks of peginterferon/ribavirin compared with HCV genotype 1 or HCV genotype 2. The aim of this study was to investigate the virological response during treatment among these three groups.
Material and MethodsStudy design and participantsThis retrospective cohort study was approved by the Ethics Committee of Mackay Memorial Hospital in Taitung’ Taiwan. We collected data from patients with chronic hepatitis C who were treated at hepatology clinics at Taitung Mackay Memorial Hospital from November 2009 to April 2012. These patients were all treated according to Taiwan National Health Insurance clinical practice guidelines (seropositive for HCV antibodies and HCV RNA, and serum alanine-imunotransferase, or ALT, above 40 IU/L). Patients were eligible for the study if they were 20 years of age or older and had naïve chronic HCV infection treated with peginterferon alpha-2b and ribavirin. The recruited patients had one of the following genotypes: genotype 1, genotype 2, and mixed genotype 1 + 2. The treatment course was 24 weeks of peginterferon alpha-2b (1.5 ug/kg/week) plus weight-based ribavirin (800 mg for those weighing < 65 kg; 1,000 mg for those weighing 65 to 80 kg; and 1,200 mg for patients weighing > 80 kg). The treatment duration was according to the Bureau of National Health Insurance in Taiwan which is reimbursing HCV treatment using 24-week combination therapy for all genotypes. The dosages of peginterferon and ribavirin were modified according to the guidelines provided by the drugs’ manufacturers.
Assessment of efficacyA rapid virological response (RVR) was defined as failure to detect HCV RNA at 4 weeks of treatment using a sensitive polymerase chain reaction (PCR)-based quantitative analysis. An early virological response (EVR) was defined as undetectable HCV RNA or at least a 2-log (10) decrease from baseline level of the serum HCV RNA at week 12 of treatment. An end of treatment virological response (EOT-R) was defined as undetectable HCV RNA at the end of 24 weeks of treatment. A sustained virological response (SVR) was defined as serum HCV RNA being undetectable 24 weeks after cessation of treatment. Those who did not have available data to assess SVR were considered to have failed to achieve SVR.
Laboratory tests/Quantitative and genotyping tests of HCV RNAAll treated patients had biweekly outpatient visits during the first month, and then monthly visits during the rest of the treatment and follow-up period. Clinical socio-demographic information, including age, gender and weight, was collected, and a medical history that included accompanying disease and any treatment side effects, was taken. Blood biochemistry was performed at enrollment included serum anti-HCV, serum HCV RNA, hepatitis B virus surface antigen (HBs-Ag), hemoglobin, aspartate aminotransferase (AST), ALT, and alpha-fetoprotein (AFP), as well as white blood cell and platelet counts. To test for liver tumor and cirrhosis, abdominal ultrasound was performed for all patients every 6 months. HCV RNA was measured by Roche COBAS®Amplicor PCR assay (Roche Molecular Diagnostics, Basel, Switzerland), with the lower limit of detection and quantitation of 15 IU/mL. HCV genotyping was performed by primer-specific PCR as previously described by Simmonds.11 The genotyping was further confirmed by direct PCR deep-sequencing using an ABI 3730 sequencer (CD Genomics, Shirley, NY, USA).
Statistical analysisAll patients who received at least one dose of medication were included in all efficacy analyses. The data of categorical variables were compared using chi-square test including odds ratios (ORs) and respective 95% confidence intervals for the association of SVR with each category of patient’s characteristics; and continuous variables were compared among the three groups using one-way analysis of variance (ANOVA). The statistical analyses were performed with an SPSS 12.0 statistical package (SPSS, Chicago, IL, USA). Missing data were treated by using the SPSS missing value options. All statistical analyses were based on two-sided hypothesis tests with a significance level of p < 0.05.
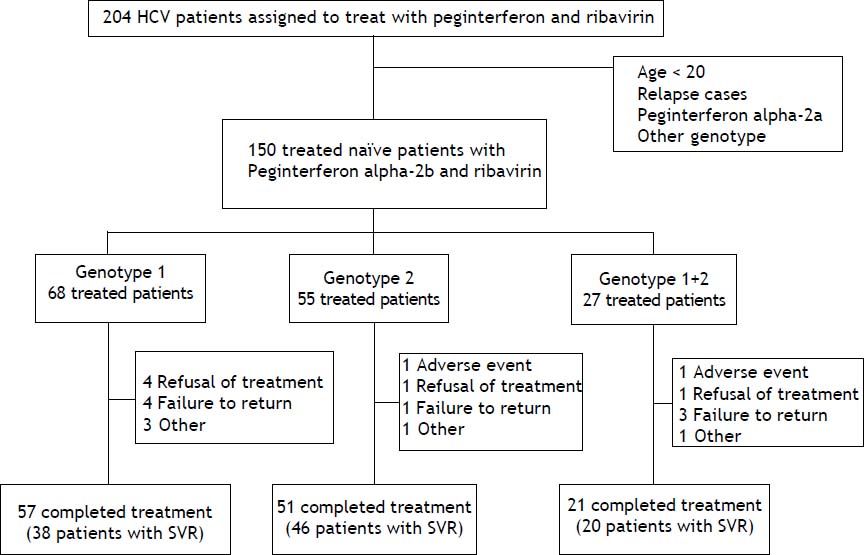
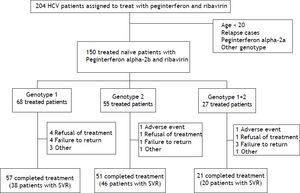
ResultsThere were 204 HCV patients assigned to treat with peginterferon and ribavirin, the percentage of HCV genotypes and subtypes analysis were genotype 1 (48.1%), genotype 2 (32.3%), genotype 1 + 2 (15.6%), genotype 6 (3.4%), and unclassified (0.5%), respectively. Those patients whose age below 20, HCV treatment relapse cases, drug with peginterferon alfa-2a, genotype 6 or unclassified were excluded. There were 150 naïve, adult chronic hepatitis C patients treated with peginterferon alpha-2b plus ribavirin included in this study. These patients were divided into 3 groups by HCV genotypes (genotype 1, genotype 2 and mixed genotype 1 + 2). The completion rate of treatment in those with genotype 1 was 83.8%, compared with 92.7% for those with genotype 2 and 77.8% for those with mixed genotype 1 + 2 (Figure 1). The average completion of treatment rate was 85%.
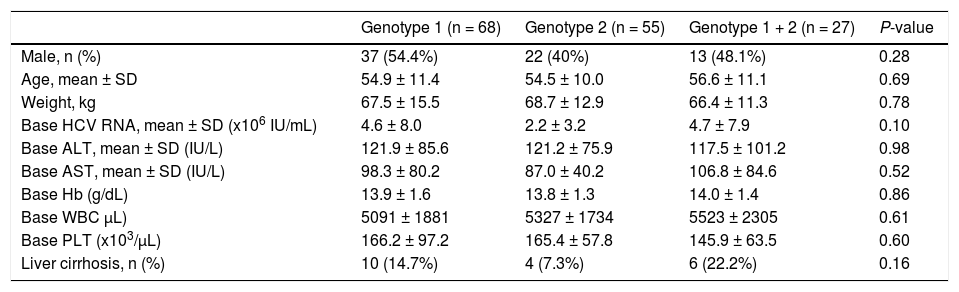
The three study groups had similar demographic and clinical characteristics; including gender, mean age, mean body weight, baseline hemoglobin levels, and baseline white blood cell and platelet counts (Table 1). The baseline mean HCV RNA serum level was high in these three groups: 2.2 ~ 4.7 x 106 IU/ mL. There were no significant differences among the 3 groups in baseline ALT and AST. The percentage of liver cirrhosis in the mixed genotype 1 + 2 group (22%) was higher than among that in the genotype 1 group (14.7%) and the genotype 2 group (7.3%), but this was not statistically significant (p = 0.16).
Demographic and clinical characters of naïve HCV treatment patients according to genotype.
| Genotype 1 (n = 68) | Genotype 2 (n = 55) | Genotype 1 + 2 (n = 27) | P-value | |
|---|---|---|---|---|
| Male, n (%) | 37 (54.4%) | 22 (40%) | 13 (48.1%) | 0.28 |
| Age, mean ± SD | 54.9 ± 11.4 | 54.5 ± 10.0 | 56.6 ± 11.1 | 0.69 |
| Weight, kg | 67.5 ± 15.5 | 68.7 ± 12.9 | 66.4 ± 11.3 | 0.78 |
| Base HCV RNA, mean ± SD (x106 IU/mL) | 4.6 ± 8.0 | 2.2 ± 3.2 | 4.7 ± 7.9 | 0.10 |
| Base ALT, mean ± SD (IU/L) | 121.9 ± 85.6 | 121.2 ± 75.9 | 117.5 ± 101.2 | 0.98 |
| Base AST, mean ± SD (IU/L) | 98.3 ± 80.2 | 87.0 ± 40.2 | 106.8 ± 84.6 | 0.52 |
| Base Hb (g/dL) | 13.9 ± 1.6 | 13.8 ± 1.3 | 14.0 ± 1.4 | 0.86 |
| Base WBC μL) | 5091 ± 1881 | 5327 ± 1734 | 5523 ± 2305 | 0.61 |
| Base PLT (x103/μL) | 166.2 ± 97.2 | 165.4 ± 57.8 | 145.9 ± 63.5 | 0.60 |
| Liver cirrhosis, n (%) | 10 (14.7%) | 4 (7.3%) | 6 (22.2%) | 0.16 |
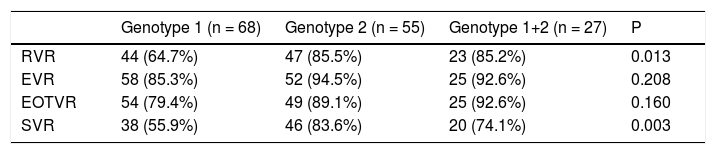
In contrast, there were significant differences in RVR rate among the 3 groups (p = 0.013). The RVR rate in the genotype 1 group was 64.7%, 85.5% in the genotype 2 group, and 85.2% in the mixed genotype 1 + 2 group. The sustained virological response (SVR) rate was 55.9% in the genotype 1 group, 83.6% in the genotype 2 group, and 74.1% in the mixed genotype 1 + 2 group. There was a significant statistical difference in SVR rate between genotype 1 and genotype 2 groups (p = 0.001). The SVR rate of mixed genotype 1 + 2 (74%) fell between that of the patients in genotype 1 and genotype 2 groups (Table 2, Figure 2).
Virological response rate according to patients who receive at least one dose of the study medication.
| Genotype 1 (n = 68) | Genotype 2 (n = 55) | Genotype 1+2 (n = 27) | P | |
|---|---|---|---|---|
| RVR | 44 (64.7%) | 47 (85.5%) | 23 (85.2%) | 0.013 |
| EVR | 58 (85.3%) | 52 (94.5%) | 25 (92.6%) | 0.208 |
| EOTVR | 54 (79.4%) | 49 (89.1%) | 25 (92.6%) | 0.160 |
| SVR | 38 (55.9%) | 46 (83.6%) | 20 (74.1%) | 0.003 |
RVR: rapid virological response. EVR: early virological response. EOTVR: end of treatment virological response. SVR: sustained virological response. P: p value.
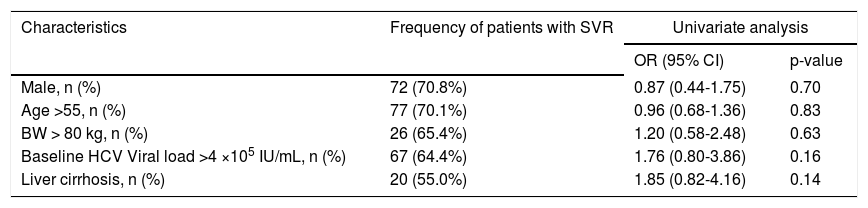
Regarding results of virological response, the sustained virological response (SVR) rate according to patients who received at least one dose of the study medication, the odds ratios according to patient’s characters, there was no significant difference (Table 3).
Sustained virological response rate and odds ratio according to patients who received at least one dose of the study medication.
| Characteristics | Frequency of patients with SVR | Univariate analysis | |
|---|---|---|---|
| OR (95% CI) | p-value | ||
| Male, n (%) | 72 (70.8%) | 0.87 (0.44-1.75) | 0.70 |
| Age >55, n (%) | 77 (70.1%) | 0.96 (0.68-1.36) | 0.83 |
| BW > 80 kg, n (%) | 26 (65.4%) | 1.20 (0.58-2.48) | 0.63 |
| Baseline HCV Viral load >4 ×105 IU/mL, n (%) | 67 (64.4%) | 1.76 (0.80-3.86) | 0.16 |
| Liver cirrhosis, n (%) | 20 (55.0%) | 1.85 (0.82-4.16) | 0.14 |
n (%): patient number and SVR percentage. OR: odds ratio.
According to previous literature, the single nucleotide polymorphisms near the interleukin-28B (IL28B) gene is significantly associated with response to Peg-IFN and ribavirin for patients with chronic genotype 1 HCV infection.12 These variations in the IL28B gene correlated with HCV viral clearance and SVR which may explain the different SVR rates among Asians, Europeans and African-Americans. Asian patients had the highest frequency of the advantageous allele in the gene for IL28B, with up to 88% of Taiwanese patients with chronic hepatitis C having the rs8099917 TT genotype which has been shown to be an important predictor of response.13 Recently, the role of IL28B genotyping is particularly relevant as direct-acting antiviral agents (DAA) are more expensive and carry a higher risk for anemia and other drug-related side effects, while decision making based on IL28B genotype may permit non-DAA-based treatment algorithms.14
Blatt, et al. reported that patients infected with HCV genotype 1 and mixed genotypes had significantly higher serum HCV RNA concentrations when compared with patients with genotype 2 and 3.15 In our study, the baseline HCV RNA serum level of patients with mixed genotype 1 + 2 was higher than that of patients with genotype 2, but this was not statistically significant. The percentage of liver cirrhosis in the mixed HCV genotype 1 + 2 group was about 22% that was higher than in the genotype 2 group (7.3%); although the difference is not statistically significant, but it should be appropriately valued and considering that mixed HCV genotype infection might have an increased prevalence of liver cirrhosis.
Currently the standard treatment course for chronic hepatitis C is 48 weeks of pegylated interferon, with the addition of ribavirin for HCV genotype 1, and 24 weeks of treatment for patients with HCV genotype 2.16 Yu, et al. reported that the SVR rate in naïve Taiwanese patients treated with 24-week course peginterferon/ribavirin was around 50%, and 90% in HCV genotypes 1 and 2 patients.17 In this study, we found that the sustained virological response rate among patients who received at least one dose of the study medication was 74.1% among those in the mixed genotype 1 + 2 group, 55.9% in the genotype 1 group, and 83.6% in the genotype 2 group. However, in the best scenario, among patients who completed treatment, the SVR rate of the mixed genotype 1 + 2 group (95.2%) was similar to that of the genotype 2 group (90.2%). Which dominant variant in HCV mixed genotype 1 + 2 is worth evaluating, since the viral dynamics are different between genotype 1 and genotype 2?18
In Taiwan, both 24 and 48 weeks of dual therapy can achieve a high SVR rate (> 96%) in patients with HCV genotype 1, with low viral load and an RVR,19 so optimizing HCV therapy in patients with HCV genotype 1 in some endemic areas is important.20 Determining the optimal treatment course for patients with mixed genotype infections is also an important issue and must take into consideration different subtypes in different geographic areas. In this study, we used pegylated interferon alpha-2b with the addition of ribavirin for 24 weeks to treat mixed HCV genotype 1 + 2 infections, and as a result achieved a 74.1% SVR rate.
Real-world data from HCV treatment in Australia reported that only 75% (414/550) of patients completed their treatment, 6.4% because of an adverse event. For 4.7%, the reason given for stopping treatment was “the patient’s decision”.21 In our study, the completion rate was 85%. Devising strategies to increase the treatment completion rate is very important, and includes developing infrastructure support and expanding treatment services to the broader HCV-infected community.
Our study had numerous limitations. First, it was a retrospective cohort study and included only a small sample of patients with mixed genotype 1 + 2 infections. Second, tests for IL-28B genotype are not currently available under Taiwanese national health insurance. Having an IL-28B genotype could predict a pretreatment response and an SVR to an abbreviated 24-week course of peginterferon/ribavirin in HCV-1 patients who did not achieve RVR and had high baseline viral loads.22
ConclusionsUsing peginterferon alpha-2b plus ribavirin for 24 weeks to treat mixed patients with HCV genotype 1 + 2 achieved a 74.1% SVR rate; this treatment efficacy was not inferior to patients with HCV genotype 1.
FundingMackay Memorial Hospital, Taitung, Taiwan.
Competing InterestsThe authors declare that they have no competing interests.
Authors’ ContributionsChing-Chung Lin, Shen-Yung Wang and Ming-Jong Bair constructed the study design, performed the data analysis, and wrote the article.
Chia-Hsien Wu, Huan-Lin Chen, I-Tsung Lin, Tsang-En Wang, Horng-Yuan Wang, Shou-Chuan Shih participated in the study design and literature review and revision of the manuscript.
All authors read and approved the final manuscript.