Chronic hepatitis D infection contributes substantially to the progression of chronic liver disease, especially in most low and middle-income countries, where hepatitis B virus-related chronic liver disease is endemic. Therefore, this study aimed to determine the magnitude and genotype of hepatitis delta virus (HDV) among patients with chronic hepatitis B (CHB)-related liver diseases in Ethiopia.
Patients and MethodsIn this cross-sectional study, 323 known HBsAg positive individuals comprising 220 patients with CHB-related liver diseases [121 advanced liver diseases (hepatocellular carcinoma /HCC/ and non-HCC) and 99 chronic hepatitis (CH)], and 103 symptomless blood donors (BD) were enrolled. An ELISA kit was employed to determine HDV infection, and quantitative real-time PCR was used to detect HDV RNA. In addition, a non-coding genomic RNA region was sequenced for genotyping and phylogenetic analysis.
ResultsIrrespective of the stage of liver disease, the overall magnitude of HDV was 7.7% (25/323). The frequency of anti-HDV increases with the severity of liver disease, 1.9%, 4%, 10%, and 21.3% among BD, CH, non-HCC, and HCC patients, respectively. HDV RNA has been detected in 1.54 %(5/323) cases with a mean viral load of 4,010,360 IU/ml. All isolates were found to be HDV genotype 1.
ConclusionsThe magnitude of HDV infection increased with the severity of liver disease, indicating HDV infection is more common among patients with CHB-related liver diseases in Ethiopia.
Hepatitis delta virus (HDV) is a satellite virus containing a negative-sense single-stranded circular RNA and the delta antigen. The virus has no established viral family but belongs to the genus Deltaviridae. Because HDV is defective, it requires the help of HBV surface antigen (HBsAg) in the course of its infection, replication, and transmission [1,2]. Thus, the prevalence of HDV infection parallels HBV distribution. Earlier reports indicated that HDV affects about 15-20 million chronic carriers of HBV worldwide [2]. Another recent pertinent report revealed a much higher prevalence with 74 million infections [3]. However, the exact estimate of hepatitis D prevalence is controversial [4].
Hepatitis D virus has similar modes of transmission as HBV and HCV, chiefly through parenteral exposure to HDV-infected individuals' blood [5]. However, mostly, the clinical course of HDV infection is more severe than that of hepatitis B and C infections. HDV can cause either acute infections (simultaneous infection with HBV, co-infection) or chronic infection (superinfection of hepatitis B infected patients) [6]. However, more than 90% of acute infections resolve spontaneously in the natural course of HDV infections, while chronic hepatitis delta infections are generally associated with a more severe and progressive form of liver disease than chronic HBV mono-infection [7,8]. Moreover, there is no protective vaccine or a well-established antiviral treatment against hepatitis D superinfection [9].
Besides its co-existence with HBV and HCV, the genotypes of HDV, as an essential viral factor, determine the progression and severity of chronic hepatitis D infection. Currently, there are eight genotypes of HDV designated as genotypes 1- 8, with sequence similarities of 60 -70 % between them [2,10]. Genotypes 1, 2, and 3 are the commonly encountered genotypes, with a worldwide distribution of genotype 1 [7,11]. Genotypes 1 and 3 are associated with more severe liver disease, whereas genotype 2 is associated with milder disease [12]. However, the pathogenicity and clinical consequences of the African clades (clades 5-8) are not well-studied [10].
In Ethiopia, various health institution-based studies implicate HBV and HCV as essential contributors to chronic liver disease [13–15]. However, information regarding the frequency and contribution of chronic hepatitis D infection to the burden of chronic liver disease is scarce. Thus, we designed this study to provide up-to-date information regarding the magnitude and circulating genotype of HDV and its contribution to the burden of chronic liver disease among CHB patients with different liver disease stages in Ethiopia.
2Materials and methods2.1Study settings and periodA health institution-based cross-sectional study was conducted in the gastroenterology (GI) clinics of three government (Tikur Anbessa Specialized Hospital, St. Paul's Hospital, and Armed Forces Hospital) and one non-government (MyungSung Christian Medical Center (MCM) /Korean Hospital/) hospitals, one specialized private clinic (Yanet specialized clinics), one medical center (Adera Medical center), and two blood banks (the National blood bank in Addis Ababa and the regional blood bank in Hawassa) from Dec. 2018 to Mar. 2019.
All study sites (the hospitals, the specialized clinic, and the Medical center) were purposefully selected for this study based on the population they serve and the availability of well-organized gastroenterology units with the proper screening and diagnostic facilities.
The study involved 323 HBsAg positive study participants, of which 220 were patients with CHB infection-related liver diseases and 103 were blood donors. The chronic liver disease (CLD) group comprises 121 advanced liver disease (AdLD) patients [hepatocellular carcinoma (HCC) (n = 61) and non-HCC (n = 60)] and 99 chronic hepatitis (CH) patients. Advanced liver disease patients were those chronic liver disease patients with HCC, advanced liver cirrhosis, fibrosis, and other related symptoms, such as jaundice and ascites, detected during a clinical examination. Patients were diagnosed based on clinicopathological analysis, biochemical (live enzyme, alpha-fetoprotein /AFP/), and imaging (Computerized tomography (CT)/ Magnetic resonance imaging (MRI)/Ultrasound (US)) modalities following the routine clinical practice in each study site. In addition, esophageal varices and fibrosis were diagnosed based on upper gastrointestinal endoscopy and Fibroscan®. A Fibroscan® level of >9.9 KPa was considered an advanced CLD. Patients with clinical (imaging) diagnosis of ascites, Esophageal varices were diagnosed as decompensated advanced CLD.
2.2Data collectionAt each study site, trained nurses collected sociodemographic characteristics and relevant clinical data, including a history of jaundice, blood transfusion, family history of HCC, and exposure to risk factors for HDV infections. Further, 10 ml of venous blood was collected from each subject and transported on dry ice to the Armauer Hansen Research Institute (AHRI) and processed. There, 4-5 ml of plasma was separated within 1-2 h of collection and held in two aliquots (1-2 ml each). The aliquots were stored at −20 °C and −70 °C until used for HDV serology and molecular analysis, respectively.
2.2.1HDV serologyThree hundred and twenty-three plasma samples positive for HBsAg were screened for HDV total antibodies (IgG and IgM) using a commercially available enzyme immunoassay (Eti-AB-Deltak-2 ELISA kit) (DiaSorin, Italy). The assay was performed as described previously [16].
2.2.2HDV detection and sequencingHDV RNA was extracted from 200 µl of plasma using a MagNA pure 96 DNA and viral nucleic acid small volume extraction kit following the manufacturer's instruction (Roche Diagnostics, Mannheim, Germany). Accordingly, RNA was successfully detected in five out of 25 anti-HDV-positive samples. The quantification was done via the Light cycler 2.0 instruments (Roche Diagnostics, Mannheim, Germany). The melting curve analysis's real-time PCR reagent concentrations, primers used, and RT conditions have been detailed before [16].
A fragment of the non-coding genome region spanning 467–834 nucleotides commonly used for HDV genotyping [17] was reverse transcribed to cDNA using the aforementioned RT-PCR parameters [16]. The PCR was performed with 5 μl of cDNA mixed with 10 μM of forward and reverse primers, 10 mM dNTP mix, 50 mM MgCl2, 5 U/μl Taq DNA polymerase (Promega, Madison, WI, USA) and 10 × buffer to a final volume of 50 μl. The PCR was performed at 94 °C for 1.5 min followed by 35 cycles at 94 °C for 30 s, 55 °C for 30 s and 72 °C for 1.5 min and a final extension for 3 min at 72 °C. The primers used were forward; 5′-AGT GAG GCT TAT CCC GG-3′ (467-483) and reverse, 5′-CTC GGA TGG CTA AGG GAG-3′ (817-834) with the nucleotides co-ordinates based on the reference sequence of AJ307077. The amplified PCR products were separated and analyzed by employing a 1.5% gel electrophoresis. Then, the Wizard® SV Gel and PCR Clean-Up System (Promega, Mannheim, Germany) were applied to purify the PCR products.
The purified PCR products were sequenced using the amplification primers by the BigDye® Terminator Cycle Sequencing Ready Reaction Kit (PE Applied Biosystems, Inc., Foster, City, CA, USA) via automated DNA sequencer, ABI Prism 3500 Genetic Analyzer (Applied Biosystems, USA).
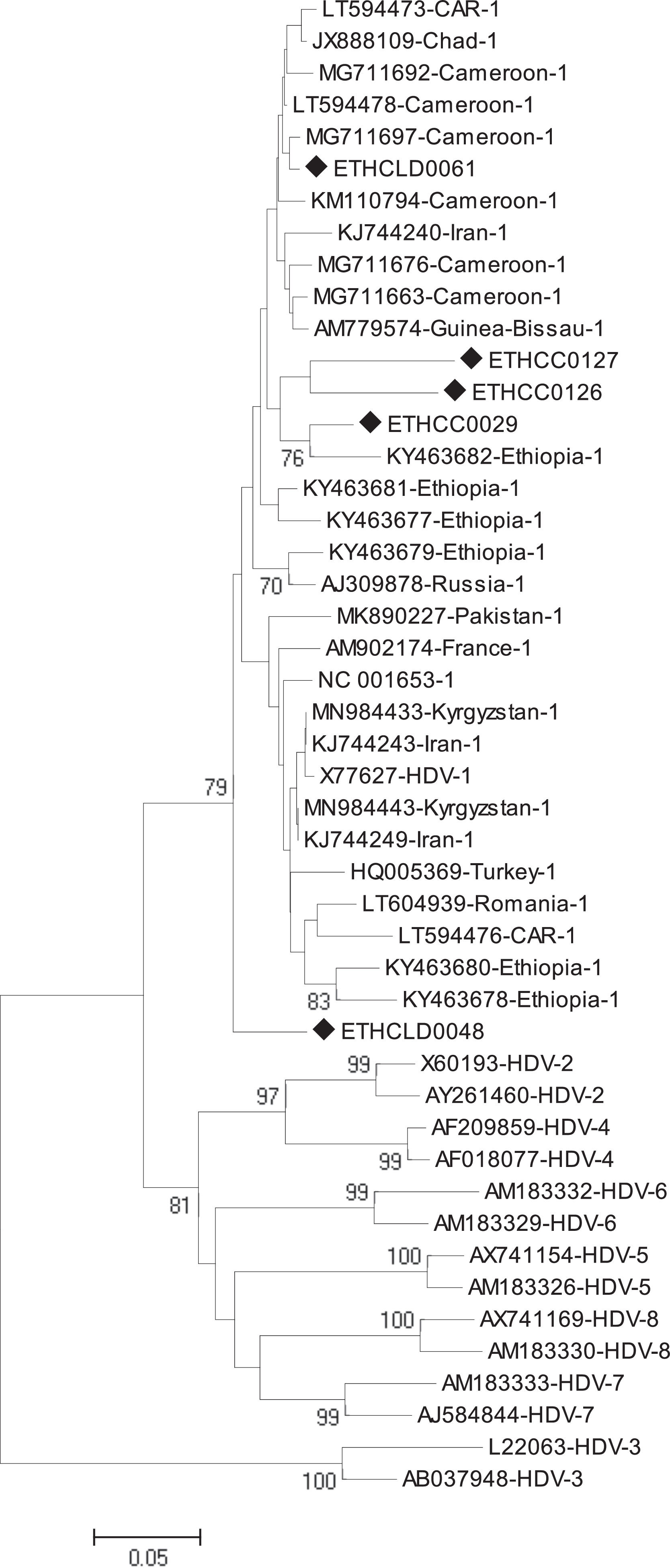
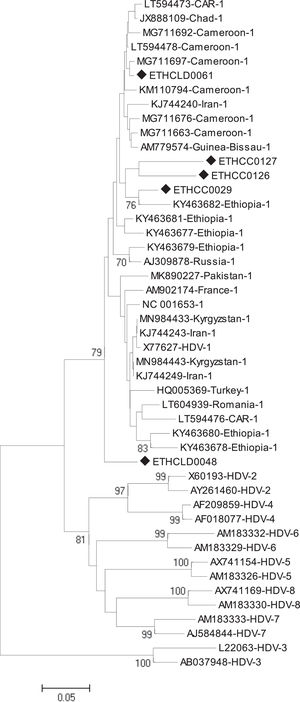
2.2.3Phylogenetic and sequence analysisThe nucleotide sequences obtained in this study were manually aligned and edited using Geneious software version 11.0.4 (http://www.geneious.com). For an independent determination of HDV genotype, the short fragment (368bp) non-coding region of the HDV genome was aligned against known sequences that represent all HDV genotypes (1 up to 8) retrieved from the National Center for Biotechnology Information (NCBI) GenBank. The evolutionary history was inferred using the Neighbor-Joining method [18]. The optimal tree with the sum of branch length = 1.85788806 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches [19]. The tree is drawn to scale, with branch lengths in the same units as the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Tamura 3-parameter method [20] and are in the units of the number of base substitutions per site. The analysis involved 47 nucleotide sequences. All positions containing gaps and missing data were eliminated. There was a total of 213 positions in the final dataset. Evolutionary analyses were conducted in MEGA6 [21].
2.3Statistical analysisThe SPSS software version 20 was used to perform all analyses in the study. Fisher's exact test and chi-square analysis were used to determine differences in demographic characteristics and potential confounders between anti-HDV positive and negative subjects and an association between HDV infection risk factors and complications of CLDs with the frequency of anti-HDV. A p-value <0.05 was considered statistically significant.
2.4Ethics statementWritten informed consent was obtained from each patient included in the study and the study protocol conforms to the ethical guidelines of the 1975 Declaration of Helsinki as reflected in a priori approval by the Ethics Committee of the College of Medicine and Health Sciences, Hawassa University (Ref. no: IRB/099/08), the College of Health Sciences, Addis Ababa University (Ref. no:056/16/DMIP), and by AHRI/ALERT ethics review committee (AAERC) (Ref. no: P025/16). In addition, permission to conduct the study was obtained from each institution involved in recruiting the study participants.
3Results3.1Sociodemographic characteristicsThe study participants' mean ± SD age was 37.59 ± 12.4, with a range of (19-83 years old). Male participants were predominant, 68%, with a male-to-female ratio of 2:1. The participants came from nearly all regions of Ethiopia, although more than half of them, 59.1%, were from the capital city, Addis Ababa (Table 1). Nearly 70% (220/323) of the study participants were CLD patients with different liver statuses. The clinical characteristics of CLD patients are summarized in Table 2.
Frequency of HDV infections by the sociodemographic characteristics of chronic liver disease patients and blood donors in Ethiopia.
CLD, Chronic liver disease; HE, Higher Education; SNNPR, Southern Nations, Nationalities, and Peoples' Region.
Clinical characteristics and laboratory profiles of patients with CHB infection-related liver diseases (N = 220).
HCC, Hepatocellular carcinoma; N-HCC, non-HCC; CH, Chronic hepatitis; AST, Aspartate aminotransferase; ALT, Alanine aminotransferase; ALP, Alkaline phosphatase; Hgb, Hemoglobuline; PT, Prothrombin Time; APTT, Activated Partial Thromboplastin Time; LFTs: Liver function tests; CLD: Chronic liver disease; HCV: Hepatitis C virus; HIV: Human Immunodeficiency virus.
A total of 323 HBsAg positive subjects, including chronic liver diseases patients (n = 220) and blood donors (n = 103), were screened for serologic evidence of HDV infection, of whom 7.7% (25/323) exhibited total antibodies against the delta antigen. The frequency of anti-HDAg antibodies was 21.3%, 10.0%, 4.0%, and 1.9% among HCC, non-HCC, chronic hepatitis patients, and blood donors, respectively.
The overall sero-distribution of anti-HDV among advanced liver disease patients was 15.7% (19/121), with a relatively higher frequency among HCC patients than non-HCC patients (21.3% vs. 10%). In addition, the frequency of anti-HDV was significantly higher in HCC patients than among blood donors (21.3% vs. 1.9%, p < 0.001).
Analysis of the magnitude of anti-HDV in relation to demographic characteristics indicated a higher anti-HDV frequency among males (8.6%), divorced (33.3%), and those aged between 38-57 years (10.42%) (Table 1). In addition, a relatively higher prevalence of anti-HDV was detected in subjects who could only read and write (lower education status) (15.4%). Moreover, the magnitude of anti-HDV exhibited a significant difference between patients from different regions, with those from the Amhara region showing the highest positivity rate (P < 0.002) (Table 1).
3.2.1Risk factors for HDV infectionThe present study assessed the frequency of exposure to risk factors of HDV infection in chronic liver disease patients (n = 220). Accordingly, 17.7% (39/220) had gum or body tattoos, 14.1% (31/220) mutilated or body pierced, 10.5% (23/220) received blood or blood products, 27.4% (60/220) visited a dental clinic, and 40.5% (89/220) undergone a major or minor surgical procedure. In addition, 39.5% (87/220) of the patients declared they had more than one sexual partner, and only 32.4% (69/213) responded that they always used condoms during sexual intercourse. Thirty-two percent (24/75) of female participants had an abortion, and of them, 62.5% (15), 16.7% (4), 12.5% (3), and 4.8% (2) declared that they had undergone an induced abortion once, two times, three times, and four and more than four times, respectively. However, none of the risk factors were significantly associated with the magnitude of HDV infection, except exposure to invasive medical procedures. That is, the frequency of anti-HDV was significantly lower in those exposed to invasive medical procedures compared with non-exposed individuals (4.5% vs. 14.5%, P < 0.02).
Furthermore, in the assessment of the association of complications of chronic liver diseases and anti-HDV status, a significantly high frequency of anti-HDV was demonstrated among patients with cirrhosis (23.0), history of jaundice (15.5%), esophageal varices (26.5%), portal hypertension (23.2%), and HCC (21.3%) than their counterparts (Table 3).
Association of the anti-HDV frequency with complications of chronic liver diseases among patients with CHB infection-related liver diseases.
CLD, Chronic liver disease; PHT, Portal hypertension; EVs, Esophageal; HCC, hepatocellular carcinoma.
Of the total of 323 HBsAg positive samples, 25 exhibited anti-HDAg antibodies. Among these, HDV-RNA was successfully detected in only 5 cases (3 HCC and 2 non-HCC). All five isolates were from advanced liver disease patients. The mean viral load of HDV was 4,010,360 IU/ml with a range of (40,800 to 11,500,000 IU/ml). The mean viral load in HCC and non-HCC patients was 6,636,667 IU/ml and 70,900 IU/ml, respectively. The phylogenetic analysis identified all five isolates as genotype 1 (HDV/I) (Fig 1). Geographically, three of the five HDV/I infected patients were from the Amhara region, and the other two were from Oromiya and Addis Ababa.
4DiscussionThe frequency of hepatitis delta virus infection, based on anti-HDV seropositivity, was 7.7%. Patients with advanced liver diseases (HCC and non-HCC) exhibited a comparatively higher frequency of anti-HDV, 15.7%. The overall magnitude of anti-HDV reported in the present study is higher than the recent hospital-based study in Addis Ababa, 1.5% [22]. Conversely, a community-based survey in Ethiopia showed a slightly higher frequency, 9.6%, of anti-HDV among subjects with previous HBV infection [23].
In this study, the frequency of anti-HDV in healthy blood donors was 1.9%, which is comparable with the previous report, 2% [24] and slightly lower than the report from northwestern Ethiopia, 3.2% [16], Cameroon, 3.4% [25], and Burkina Faso, 2.5% [26]. This implies that, despite the high overall prevalence of HDV, the cohort-specific prevalence among healthy blood donors is still low. Furthermore, the prevalence showed no change from the result reported two decades ago in Addis Ababa [24]. Nevertheless, a comprehensive nationwide study with a larger sample size is required to confirm our findings.
The cohort-specific prevalence of anti-HDV among HBsAg positive chronic hepatitis (CH) patients was 4%, which is somehow higher than the previous report from Addis Ababa, 1.5% [22]. However, in the report from Aberra et al., 12 patients exhibited active HDV infection markers, while in our study, only five patients exhibited an active infection. Thus, variation in HDV prevalence in these two studies might be due to methodological or geographic differences. As indicated in the previous study [22] and a similar study from Libya [27], “micro-epidemiology” could be one factor for the difference in the prevalence report of HDV infection between different regions of Ethiopia. For instance, in the previous study, most of the anti-HDV-positive subjects came from Addis Ababa, whereas, in the current study, the majority of them were from the Amhara region, followed by other areas different from the four (Addis Ababa, SNNPR, Amhara, and Oromia) regions.
Our study showed a moderately high anti-HDV frequency in AdLD patients, 15.7% (19/121), which is considerably lower than the report from Mongolia, where HDV is endemic. In addition, the report indicated that of 207 CLD patients of various stages, 128 (61.8%) and 117 (56.5%) exhibited anti-HDV and HDV-RNA, respectively [28]. A related study from Tajikistan also reported a comparatively higher anti-HDV level among CLD patients with cirrhosis and HCC (35.2%) [29]. However, our findings are comparable with the report from Addis Ababa, which showed a 17.2% frequency of anti-HDV in AdLD (cirrhosis and HCC) patients [24]. Furthermore, a comparable seropositivity rate of anti-HDV in HCC patients was demonstrated in the current study and the previous one in Ethiopia (21.3% and 23%) [24]. Likewise, the rate of anti-HD in non-HCC advanced liver disease patients was 10.0% (6/60), which is comparable with the report from northwestern Ethiopia, 12.7% [16] and slightly lower than the report by Tsega et al. (16%) [24]. Thus, findings from the current and previous studies suggest relatively stable transmission dynamics of the hepatitis delta virus among chronic liver disease patients in Ethiopia.
Concurrent infection with HBV and HDV accelerates chronic liver diseases' progress to the advanced stages, such as decompensated cirrhosis and HCC, than HBV mono-infection could. Miao et al. reported that the burden of Hepatitis D infection is higher among HBsAg-positive, progressive liver disease patients (with fulminant hepatitis, cirrhosis, or HCC) compared to asymptomatic carriers [29]. Likewise, in this study, the magnitude of anti-HDV across the different clinical groups was substantially different, with a higher frequency in AdLD patients (HCC and non-HCC) than in blood donors (15.7% vs. 1.9%). Implying that HDV infection is common among CHB-related liver disease patients and might contribute to the burden of liver disease in Ethiopia. Nonetheless, compared to other nations where HBV and HDV are endemic, the general and cohort-specific distribution of HDV and the infection burden is lower (e.g., in Mongolia, the prevalence of anti-HD in CLD patients reaches 80 %) [28,30].
Similar to the prior report [22], the present study also suggests geographic heterogeneity in HDV prevalence in Ethiopia, although it is difficult to conclude with such a small sample size. A related study from Libya and Kenya also demonstrated variations in HDV infections in different localities of the same country [27,31]. It is probably due to differences in the distribution of infection risk factors and the population's exposure status in different localities.
The five isolates in the current study were from patients with advanced liver disease. The significantly high mean viral load in HCC than non-HCC patients (6,636,667 IU/ml Vs. 70,900 IU/ml) might be due to the substantial role of HDV superinfection in the progression of chronic liver disease, although it is difficult again to conclude with such a small sample size. According to Romeo and Perbellini, an HDV viral load greater than 600,000 IU/ml is associated with liver disease progression to an advanced stage, such as cirrhosis [32].
Currently, eight major HDV clades are identified in various parts of the world [33,34]. The most divergent HDV strain is clade three from South America [33,34]. On the other hand, HDV/I has ubiquitous worldwide distribution [35]. In parallel, HDV/I was the only genotype identified in the present study, consistent with prior reports from Ethiopia [16,22].
As HDV/I has a worldwide distribution, the chronicity and severity of the liver disease it causes vary from place to place. For example, Wu et al. found that, compared to HDV/II, genotype 1 is more frequently associated with acute fulminant hepatitis and progression to cirrhosis and HCC in the same geographic area [36]. Thus, large-scale surveys might be required to confirm the HDV genotype distribution in Ethiopia, as evidence of the distribution of HDV types is vital in determining vaccine strategies and patient management.
Because there are minimal (2-3) molecular studies on HDV in Ethiopia, all the above information (concerning the prevalence, genotype, transmission dynamics, and geographic distribution of HDV and its impact on liver disease progression) provided by the current study is critical inputs to the prevention and management strategy of HDV-related liver diseases in Ethiopia.
However, this study also has some limitations; such as being a health institution-based study, the prevalence of HDV infection among HBsAg-positive individuals is somehow higher. Therefore, it might not represent HDV's actual prevalence in Ethiopia's general population. In addition, the small number of HDV-positive study subjects might affect our conclusion on the association between several factors and HDV infection.
5ConclusionsThe magnitude of HDV infection is high, with all clades identified being genotype 1. Besides, the prevalence of anti-HDV increases with the severity of liver disease with all the active HDV infections detected among patients with AdLD, magnifying the common occurrence of HBV/HDV co-infection among chronic liver disease patients.
Thus, we recommend screening and treating HDV infection in individuals with AdLD. In addition, nationwide studies are mandatory to cognize the actual burden of HDV and incite policy changes on prevention measures.
Declaration of interestNone.
CRediT authorship contribution statementYayehyirad Tassachew: Conceptualization, Data curation, Formal analysis, Methodology, Project administration, Writing – original draft, Writing – review & editing. Yeshambel Belyhun: Formal analysis, Methodology, Supervision, Writing – review & editing. Tamrat Abebe: Supervision, Writing – review & editing. Adane Mihret: Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing. Tezazu Teffera: Methodology, Writing – review & editing. Girma Ababi: Methodology, Writing – review & editing. Abate Shewaye: Methodology, Writing – review & editing. Hailemichael Desalegn: Methodology, Writing – review & editing. Abraham Aseffa: Funding acquisition, Supervision, Writing – review & editing. Andargachew Mulu: Project administration, Resources, Supervision, Writing – review & editing. Rawleigh Howe: Funding acquisition, Supervision, Project administration, Writing – review & editing. Uwe G. Liebert: Resources, Supervision, Writing – review & editing. Melanie Maier: Resources, Supervision, Writing – review & editing.
An indirect fund was secured for this study from Addis Ababa University, Hawassa University, and AHRI. The sponsors had no involvement in any of the stages from study design to submission of this manuscript for publication.