Long non-coding RNA (lncRNA) has been shown to be a vital regulator of cancer progression, including hepatocellular carcinoma (HCC). However, the role of DEAD/H box protein 11 antisense RNA 1 (DDX11-AS1) in HCC remains to be further studied.
Material and MethodsThe expression levels of DDX11-AS1, miR-195-5p and metastasis-associated in colon cancer-1 (MACC1) were determined by quantitative real-time PCR (qRT-PCR). Cell counting kit-8 (CCK-8), transwell and apoptosis determination assays were used to evaluate cell proliferation, migration, invasion and apoptosis, respectively. Mice xenograft models were constructed to verify the effect of DDX11-AS1 on HCC tumor growth in vivo. Furthermore, lactate production, glucose consumption, ATP level and glucose uptake were detected to assess cell glucose metabolism. The interactions among DDX11-AS1, miR-195-5p and MACC1 were verified by dual-luciferase reporter assay and RNA immunoprecipitation (RIP) assay. Moreover, western blot (WB) analysis was performed to evaluate the protein levels.
ResultsDDX11-AS1 was upregulated in HCC tissues and cells, and its silencing could inhibit HCC cell proliferation, migration, invasion and glucose metabolism, and promote apoptosis in vitro. Also, DDX11-AS1 knockdown reduced HCC tumor growth in vivo. Besides, DDX11-AS1 could interact with miR-195-5p, and miR-195-5p inhibitor reversed the inhibitory effect of silenced DDX11-AS1 on HCC cell progression. In addition, MACC1 was a target of miR-195-5p, and its overexpression reversed the suppression effect of miR-195-5p on HCC cell progression.
ConclusionOur data revealed that DDX11-AS1 could act as an oncogenic regulator in HCC, providing a potential therapeutic target for HCC treatment.
Hepatocellular carcinoma (HCC) is a common malignant tumor with high mortality mainly due to the high rate of metastasis and recurrence of HCC [1–3]. Delayed diagnosis and poor prognosis seriously hinder the cure rate of HCC [4,5], so the treatment of HCC remains challenging. Therefore, the search for effective therapeutic targets can help us provide ideas for treatment of HCC.
Long non-coding RNAs (lncRNAs) are transcripts of more than 200 nts in length [6]. Studies show that lncRNAs may act as crucial parts in many diseases and cancers [7,8]. The abnormal expression of lncRNA can affect the ability of tumor occurrence, growth and metastasis [9], suggesting that lncRNA is expected to be an effective target for tumor diagnosis and treatment. Many lncRNAs could participate in the regulation of HCC processes. For example, lncRNA AWPPH promoted HCC cell proliferation, migration, and tumor growth in vivo via regulating YBX2 [10]. LncRNA HOST2 increased the EMT, proliferation, invasion and metastasis of HCC cells by activating the JAK2/STAT3 signaling pathway [11]. Recent studies had found that lncRNA DEAD/H box protein 11 antisense RNA 1 (DDX11-AS1) was abnormally expressed in HCC [12,13]. However, few studies had been conducted on the molecular mechanisms of DDX11-AS1 in HCC development.
MicroRNAs (miRNAs) are a class of endogenous non-coding small molecules, which control gene expression through targeting mRNAs to trigger translation inhibition or RNA degradation [14,15]. Many evidences confirm that miRNAs may function as oncogenes or tumor suppressors in the development of various human cancers, including HCC [16–18]. With the deepening of studies, researchers have found that lncRNA can act as “miRNA sponge” to regulate the expression of downstream genes [19,20]. Therefore, the hypothesis of the lncRNA-miRNA-mRNA axis is an important way to elucidate the molecular mechanism of lncRNA.
The research aimed to investigate the effect and mechanism of DDX11-AS1 on HCC progression. In this study, we uncovered that DDX11-AS1 was overexpressed in HCC patients and cells. Through the loss- and gain-functional experiments, we explored the function of DDX11-AS1 on the progression of HCC. Bioinformatics prediction and further experimental investigated the mechanism of DDX11-AS1. The discovery of DDX11-AS1/miR-195-5p/MACC1 axis enriched the regulatory network of DDX11-AS1, and provided the basis for the treatment of HCC.
2Materials and methods2.1Samples collection37 paired HCC tissues and adjacent normal tissues samples used in this study were collected from 37 HCC patients in The First College of Clinical Medical Science of China Three Gorges University. According to the different stages of HCC patients, HCC tissues were divided into early stage (I + II) and late advanced stage (III + IV). All patients signed informed consent, and this study was authorized by the ethics committee of The First College of Clinical Medical Science of China Three Gorges University.
2.2Cell culture and transfectionHCC cell lines (Huh7 and PCL/PRF/5) and human hepatic cell line (THLE-2) were bought from American Type Culture Collection (ATCC, Manassas, VA, USA). All cells were cultured in DMEM (Solarbio, Beijing, China) containing 10% fetal bovine serum (FBS, Sijiqing, Hangzhou, China) and 1% penicillin/streptomycin at 37 °C in 5% CO2 incubator.
Small interfering RNA against DDX11-AS1 (si-DDX11-AS1) or overexpression vector (DDX11-AS1) and their controls (si-NC or pcDNA), miR-195-5p mimic or inhibitor (miR-195-5p or in-miR-195-5p) and their controls (miR-NC or in-miR-NC), MACC1 overexpression vector (MACC1) and its control (pcDNA) were provided by Vigene Biosciences (Shandong, China). Lentivirus harboring short hairpin RNA against DDX11-AS1 (sh-DDX11-AS1) and its control (sh-NC) were constructed by Genewiz (Suzhou, China). The transfection was executed by Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA).
2.3Quantitative real-time PCR (qRT-PCR)The RNA was extracted by TRIzol reagent (Invitrogen) and reverse-transcribed into cDNA by HiScript 1st Strand cDNA Synthesis Kit (Vazyme, Nanjing, China). QRT-PCR was executed using SYBR Green (Takara, Dalian, China). Fluorescence signals were collected and analyzed by PCR system. Relative expression was determined by 2−ΔΔCT method and normalized by 18S ribosomal RNA (rRNA) or U6. The primer sequences were listed as follows: DDX11-AS1, F 5’-CTGGCTACTCTTCCTCCTGG-3’; R 5’-CAGAGGACATGTGGGAGGTT-3’; miR-195-5p, F 5’-GAATTCGCCTCAAGAGAACAAAGTGGAG-3’; R 5’- AGATCTCCCATGGGGGCTCAGCCCCT-3’; MACC1, F 5’-TCGGTCAGGAAGAATTGCAC-3’; R 5’-TTGTGAAGCAAGTCTGGGTCC-3’; 18S rRNA, F 5’-ATCGGGGATTGCAATTATTC-3’, R 5’-CTCACTAAACCATCCAATCG-3’; U6, F 5’-CGCTTCGGCAGCACATATAC-3’; R 5’-AAATATGGAACGCTTCACGA-3’.
2.4Subcellular fractionation and localizationThe Cytoplasmic & Nuclear RNA Purification Kit (Norgen Biotek, Thorold, ON, Canada) was used to isolate the cytoplasmic and nuclear RNAs of Huh7 and PCL/PRF/5 cells. U6 and 18S rRNA were used as nuclear control and cytoplasm control, respectively. QRT-PCR was performed to assess the expression of DDX11-AS1, U6 and 18S rRNA in the cytoplasmic and nuclear of cells.
2.5Cell counting kit-8 (CCK-8) assayHuh7 and PCL/PRF/5 cells were inoculated into 96-well plates. CCK-8 Kit (Medchemexpress, Shanghai, China) was used for detecting the proliferation ability of cells at 0, 24, 48, and 72 h. The OD value was determined using a microplate reader (Molecular devices, Shanghai, China) at 450 nm.
2.6Transwell assayHuh7 and PCL/PRF/5 cells were inoculated in upper chamber of uncoated and pre-coated with Matrigel (BD Bioscience, San Jose, CA, USA) containing serum-free DMEM for the detection of cell migration and invasion, respectively. DMEM containing 10% FBS was added to the lower chamber. After 24 h, cells of the lower chamber were fixed and stained. Then, the numbers of migrated and invaded cells were counted with an inverted microscope (Leica, Weztlar, Germany) to calculate cell migration and invasion abilities.
2.7Apoptosis determinationHuh7 and PCL/PRF/5 cells were collected into a centrifuge tube after digested by trypsin. After centrifugation, cells were incubated with Annexin V-FITC + PI reagent (Beyotime, Shanghai, China) for 15 min. FITC fluorescence was detected by flow cytometry.
2.8Mice xenograft modelsThe one-month-old male BALB/c-nude mice were brought in Yison Bio (Shanghai, China). Huh7 cells transfected with sh-DDX11-AS1 or sh-NC were prepared and injected into nude mice. Tumor length and width were recorded every 7 d, and tumor volume was calculated by length × width2/2. The tumor was removed and weighted after 28 d. The animal model experiment was authorized by the animal research committee of The First College of Clinical Medical Science of China Three Gorges University.
2.9Measurement of lactate production, glucose consumption, ATP level and glucose uptakeFor detecting the lactate production and glucose consumption, the culture mediums of Huh7 and PCL/PRF/5 cells were collected. According to the manufacturer’s instructions, the lactate production and glucose consumption were detected using Lactate Assay Kit and Glucose Assay Kit (Biovision, Palo Alto, CA, USA), respectively. For measuring the ATP level, Huh7 and PCL/PRF/5 cells (1 × 104) were seeded in white opaque microplate. Then, the ATP level was determined using ATP Assay Kit (Abnova, Taibei, Taiwan) according to the manufacturer’s instructions. For testing the glucose uptake, Huh7 and PCL/PRF/5 cells were seeded in 96-well plates. After 24 h, the culture medium was removed, and 50 μL 2-deoxyglucose (2-DG) was added into cells and incubated for 20 min. The glucose uptake was detected using Glucose Uptake Assay Kit (Abnova) according to the manufacturer’s instructions.
2.10Western Blot (WB) analysisThe protein was extracted using RIPA buffer (Genetex, San Antonio, Texas, USA) and quantified using BCA Protein Assay Kit (Genetex). The same amount of protein was isolated on 10% SDS-PAGE gel and transferred to PVDF membranes. Then, the membranes were closed with 5% non-fat milk and cultured with the primary antibody against Cyclin D1 (1:1,000, CST, Boston, MA, USA), p21 (1:500, Beyotime), Bcl-2 (1:1,000, Beyotime), Bax (1:5,000, Beyotime), E-cadherin (1:1,000, CST), N-cadherin (1:1,000, CST), Vimentin (1:5,000, Beyotime), MACC1 (1:1,000, CST), β-actin (1:1,000, CST) at 4℃ overnight. Subsequently, the membranes were cultured with the horseradish peroxidase (HRP)-conjugated secondary antibody (1:1,000, CST) for 1 h. BeyoECL Star (Beyotime) was used to visualize the protein signals and Image-pro plus software 6.0 was used to analyze the results.
2.11Dual-luciferase reporter assayAccording to the binding sites of miR-195-5p with DDX11-AS1 and MACC1 3’UTR predicted by DIANA and Targetscan online software, the wild-type (WT) and mutant-type (MUT) sequences of DDX11-AS1 (WT: 5’-CCAUGGGUAUAAGUGCUGCUA-3’; MUT 5’-CCAUGGGUAUAACACGACGAU-3’) and MACC1 3’UTR (WT: 5’-CUUUUCUCCCACAUGCUGCUG-3’; MUT: 5’-CUUUUCUCCCACAACGACGAG-3’) were amplified and inserted into the psi-CHECK-2 reporter vector (Promega, Madison, WI, USA), respectively, which were synthesized by General Biosystems (Anhui, China). MiR-195-5p mimic and miR-NC were co-transfected with the above vectors into Huh7 and PCL/PRF/5 cells. After 48 h, the relative luciferase activity (Firefly/Renilla) was measured by Dual-Luciferase Reporter Assay System (Beyotime) according to the manufacturer’s instructions.
2.12RNA immunoprecipitation (RIP) assayThis assay was performed by RIP Kit (Millipore, Billerica, MA, USA). After transfected miR-195-5p mimic or miR-NC, Huh7 and PCL/PRF/5 cells were lysed by RIP lysis buffer and incubated with magnetic beads coupled with Ago2 (RIP-Ago2) or IgG (RIP-IgG). The enrichments of DDX11-AS1 and MACC1 were analyzed by qRT-PCR.
2.13Statistical analysisData were expressed as mean ± SD. All date were performed by Student’s t-test and one-way ANOVA. The differences among groups were compared by SPSS v.16.0 (SPSS, Inc., Chicago, IL, USA). P < 0.05 was considered to be significant.
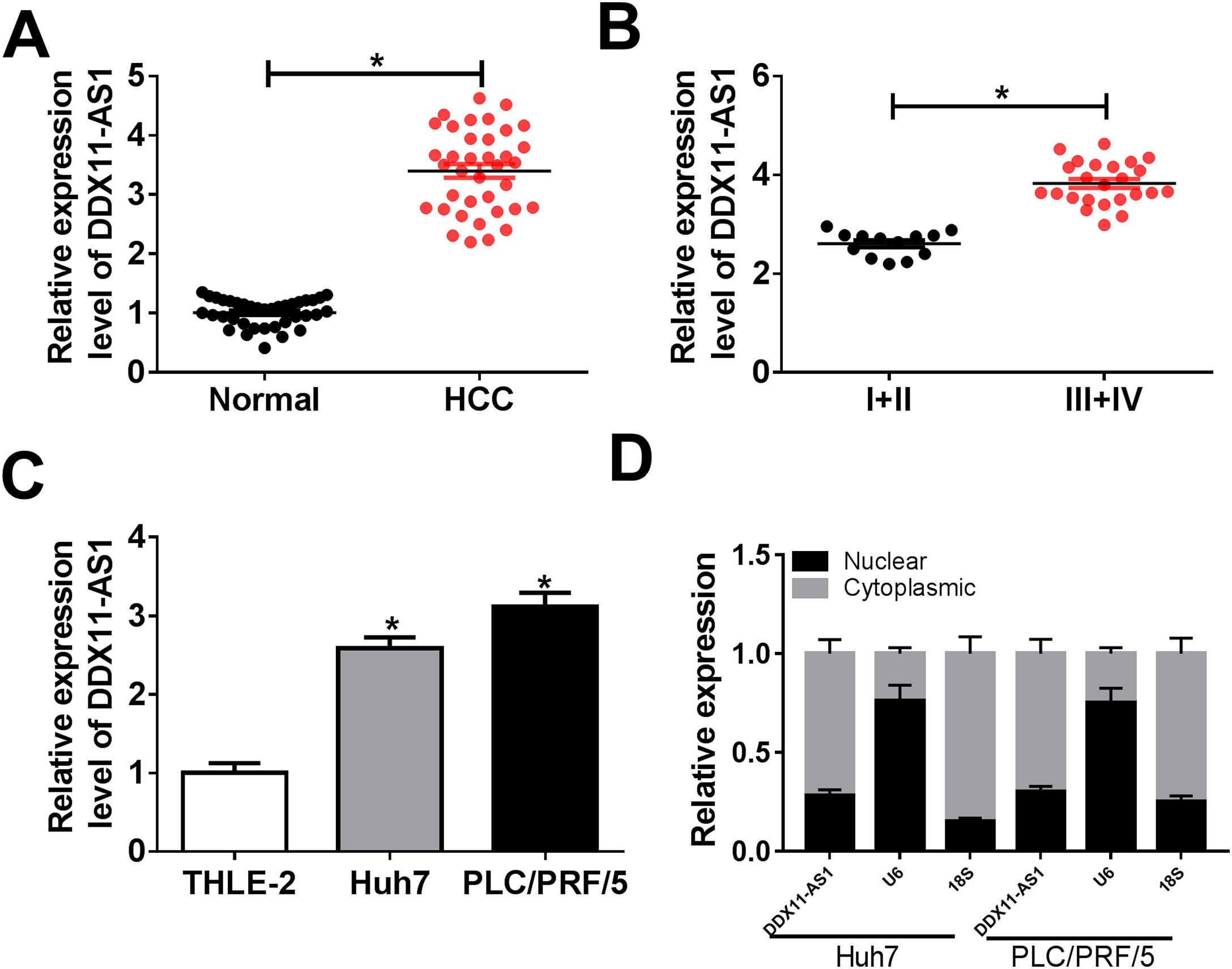
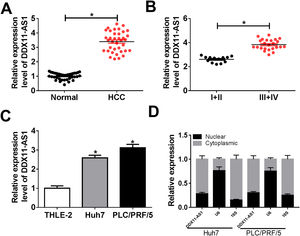
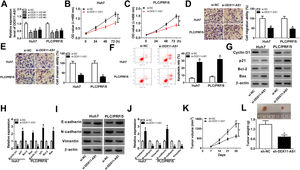
3Results3.1DDX11-AS1 was highly expressed in HCCFirstly, we examined the DDX11-AS1 expression in HCC tissues and cells. Through the detection of DDX11-AS1 expression in 37 paired HCC tissues and adjacent normal tissues, we found that DDX11-AS1 was highly expressed in HCC tissues (Fig. 1A). Depending on the stage of the HCC patients, we also discovered that DDX11-AS1 expression was significantly higher in the tissues of the advanced HCC patients (III + IV) than in the early stage (I + II) (Fig. 1B). Also, DDX11-AS1 was upregulated in HCC cells (Huh7 and PLC/PRF/5) compared with that in THLE-2 cells (Fig. 1C). Therefore, we speculated that DDX11-AS1 might play an important role in the progression of HCC. In addition, by detecting the expression of DDX11-AS1 in the nuclear and cytoplasmic of Huh7 and PLC/PRF/5 cells, we uncovered that DDX11-AS1 mainly accumulated in the cytoplasmic of HCC cells (Fig. 1D).
DDX11-AS1 was upregulated in HCC. (A) The expression of DDX11-AS1 was increased in HCC tissues compared to adjacent normal tissues. (B) QRT-PCR was used to measure DDX11-AS1 expression in early stage (I + II) and advanced stage (III + IV) of HCC patients. (C) The expression of DDX11-AS1 was evaluated by qRT-PCR in THLE-2 cells and HCC cell lines (Huh7 and PLC/PRF/5). (D) QRT-PCR was used to detect the expression of DDX11-AS1, U6 and 18S in the nuclear and cytoplasmic of Huh7 and PLC/PRF/5 cells. *P < 0.05.
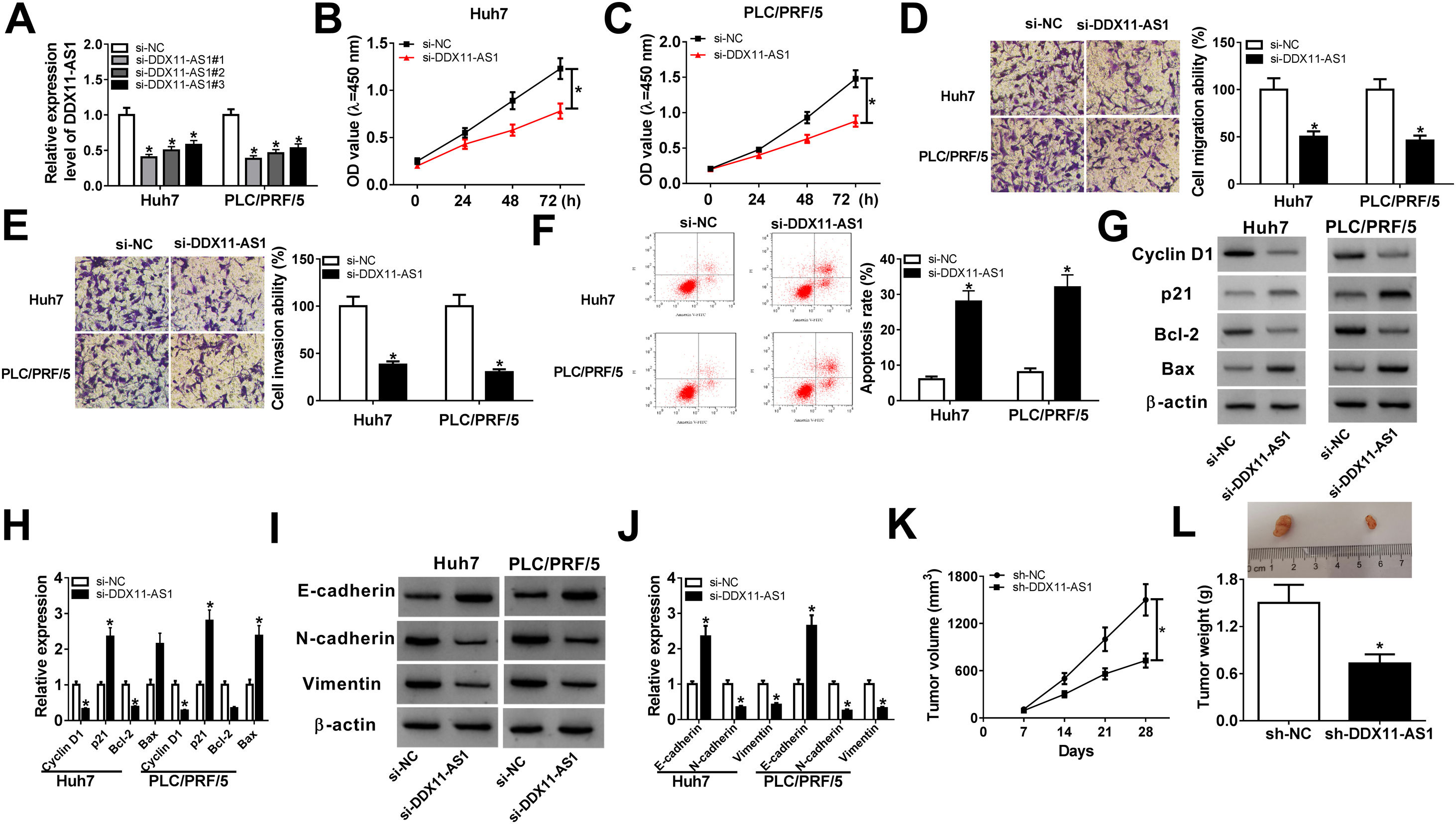
To further explored the role of DDX11-AS1 in HCC cells, we transfected three si-DDX11-AS1 into Huh7 and PLC/PRF/5 cells. As shown in Fig. 2A, all three si-DDX11-AS1 could effectively inhibit the expression of DDX11-AS1, especially si-DDX11-AS1#1. Therefore, si-DDX11-AS1#1 was used for follow-up experiments. CCK-8 assay results showed that knockdown of DDX11-AS1 decreased the proliferation abilities of Huh7 and PLC/PRF/5 cells (Fig. 2B–C). Besides, transwell assay results further demonstrated that DDX11-AS1 silencing markedly decreased the migration and invasion abilities of HCC cells compared with the negative control (Fig. 2D–E). Moreover, cell apoptosis was examined by apoptosis determination assay, and the results indicated that DDX11-AS1 silencing obviously increased the apoptosis rates of Huh7 and PLC/PRF/5 cells (Fig. 2F). Through measuring the protein levels of cell cycle-related markers (Cyclin D1 and p21), apoptosis-related markers (Bcl-2 and Bax) and metastasis-related markers (E-cadherin, N-cadherin and Vimentin), we found that silenced DDX11-AS1 reduced the protein levels of Cyclin D1, Bcl-2, N-cadherin and Vimentin, while increased the protein levels of p21, Bax and E-cadherin (Fig. 2G–J). These indicated that DDX11-AS1 regulated the proliferation, metastasis and apoptosis of HCC by mediating cell cycle, epithelial-mesenchymal transition and apoptosis markers expression. Additionally, HCC mice xenograft models were constructed to verify the effect of DDX11-AS1 on HCC tumors in vivo. As presented in Fig. 2K–L, we observed that DDX11-AS1 interference could remarkably reduce HCC tumor volume and tumor weight. These results indicated the DDX11-AS1 might have a pro-cancer role in HCC.
DDX11-AS1 silencing suppressed HCC cell progression and HCC tumor growth. (A) The transfection efficiency of si-DDX11-AS1#1/#2/#3 was determined by qRT-PCR assay. (BC) CCK-8 assay was used to measure the proliferation of Huh7 and PLC/PRF/5 cells. (DE) Transwell assay was performed to detect the migration and invasion of Huh7 and PLC/PRF/5 cells. (F) The apoptosis rate of Huh7 and PLC/PRF/5 cells was determined by apoptosis determination assay. (GJ) The protein levels of Cyclin D1, p21, Bcl-2, Bax, E-cadherin, N-cadherin and Vimentin were determined using WB analysis. Tumor volume (K) and tumor weight (L) of HCC mice models were detected in the sh-DDX11-AS1 group and sh-NC group. *P < 0.05.
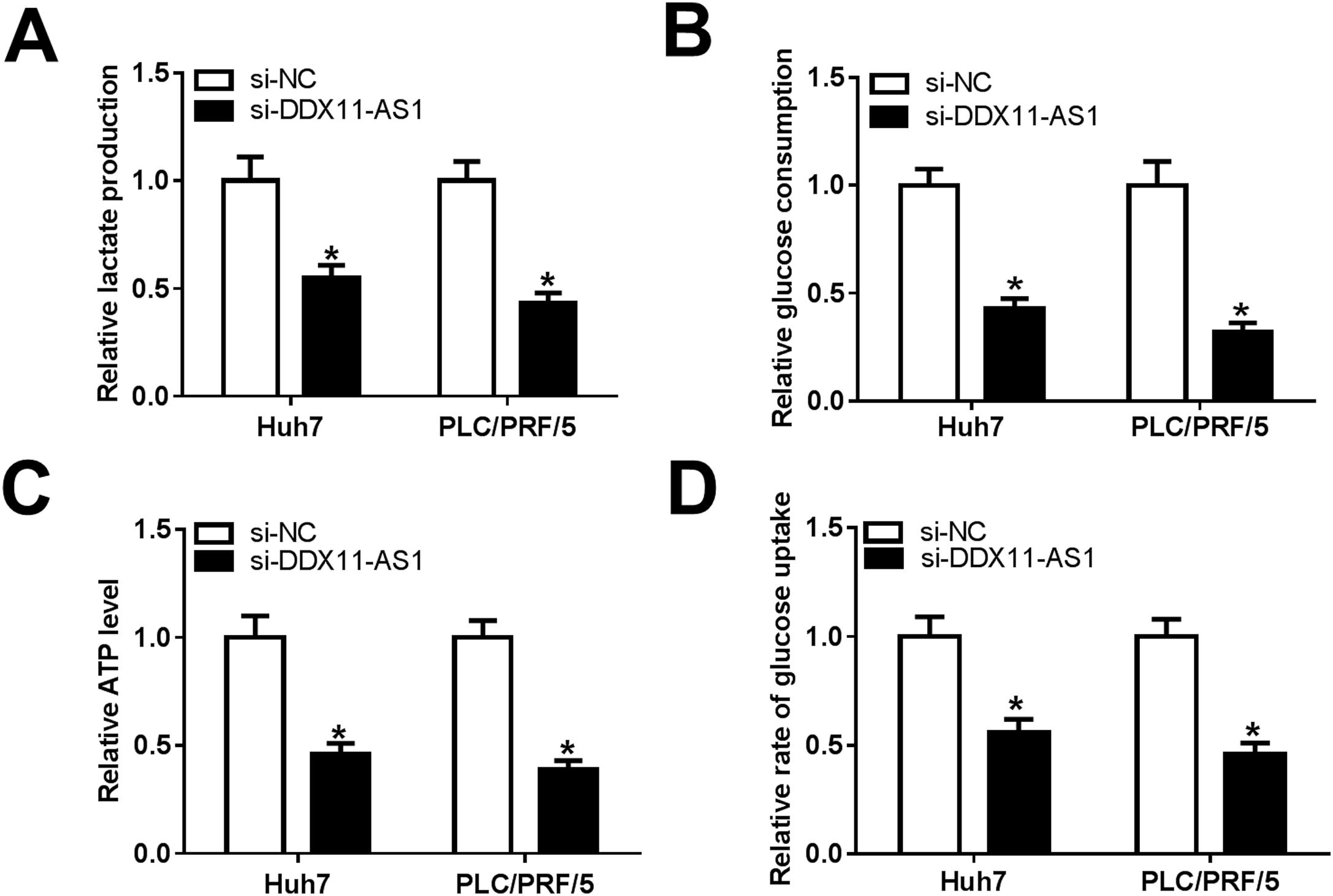
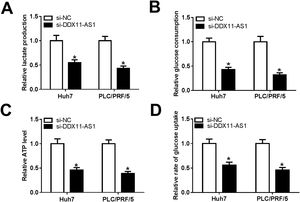
Glucose metabolism is an important source of energy during the growth and migration of cancer cells. Here, we also evaluated the glucose metabolism of HCC cells. By detecting the lactate production and glucose consumption of cells, we found that DDX11-AS1 silencing could repress the lactate production and glucose consumption of Huh7 and PLC/PRF/5 cells (Fig. 3A–B). Additionally, the ATP level and glucose uptake of HCC cells also could be inhibited by DDX11-AS1 knockdown (Fig. 3C–D). All data indicated that downregulation of DDX11-AS1 could hinder the glucose metabolism of HCC cells.
DDX11-AS1 knockdown inhibited the glucose metabolism of HCC cells. The lactate production (A), glucose consumption (B), ATP level (C) and glucose uptake (D) of Huh7 and PLC/PRF/5 cells were detected by Lactate Assay Kit, Glucose Assay Kit, ATP Assay Kit and Glucose Uptake Assay Kit, respectively. *P < 0.05.
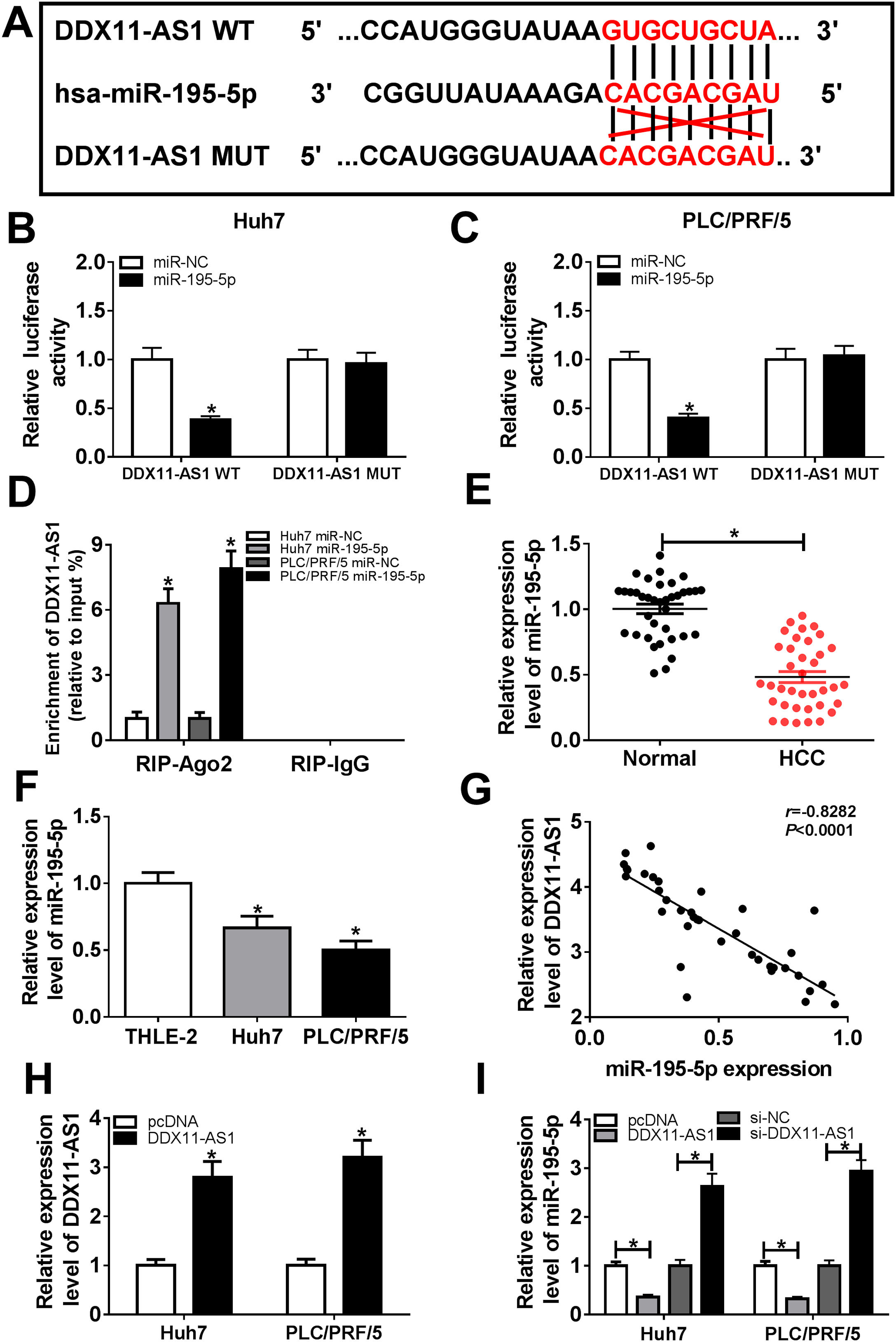
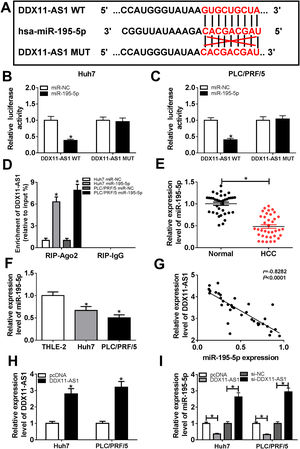
In order to understand the mechanism of DDX11-AS1, the DIANA tool was performed to predict the miRNAs that could bind with DDX11-AS1, and the results disclosed that DDX11-AS1 had binding sites with miR-195-5p (Fig. 4A). Dual-luciferase reporter assay showed that miR-195-5p mimic markedly decreased the luciferase activity of DDX11-AS1 WT reporter vector, while had no influence on that of DDX11-AS1 MUT reporter vector in Huh7 and PLC/PRF/5 cells (Fig. 4B–C). Also, RIP assay results indicated that miR-195-5p mimic remarkably enhanced the enrichment of DDX11-AS1 in RIP-Ago2 complex (Fig. 4D). Besides, we measured the expression of miR-195-5p in HCC tissues and cells, and found that miR-195-5p was markedly decreased in HCC tissues and cells compared with that in matched negative controls (Fig. 4E–F). Subsequently, correlation analysis showed that miR-195-5p expression was negatively correlated with DDX11-AS1 expression in HCC tissues (Fig. 4G). To further verify the effect of DDX11-AS1 expression on miR-195-5p expression, we constructed DDX11-AS1 overexpression vector and confirmed its transfection efficiency by detecting DDX11-AS1 expression using qRT-PCR in Huh7 and PLC/PRF/5 cells (Fig. 4H). Through determining miR-195-5p expression, we discovered that miR-195-5p expression was inhibited by DDX11-AS1 overexpression, while promoted by DDX11-AS1 silencing in Huh7 and PLC/PRF/5 cells (Fig. 4I). These results showed that DDX11-AS1 could directly bind with miR-195-5p in HCC.
DDX11-AS1 could interact with miR-195-5p in HCC. (A) The predicted target sequences and mutated sequences between DDX11-AS1 and miR-195-5p were shown. (B–C) Dual-luciferase reporter assay was used to detect the interaction between DDX11-AS1 and miR-195-5p in Huh7 and PLC/PRF/5 cells. (D) RIP assay was performed to measure the enrichment of DDX11-AS1 in RIP-Ago2 and RIP-IgG. The expression of miR-195-5p in HCC tissues (E) and cells (F) was determined by qRT-PCR. (G) The correlation between DDX11-AS1 and miR-195-5p expression was determined by Pearson correlation analysis. (H) The transfection efficiency of DDX11-AS1 overexpression vector was detected by qRT-PCR. (I) The expression of miR-195-5p was tested by qRT-PCR in Huh7 and PLC/PRF/5 cells transfected DDX11-AS1 overexpression vector or si-DDX11-AS1 and their negative control (pcDNA or si-NC). *P < 0.05.
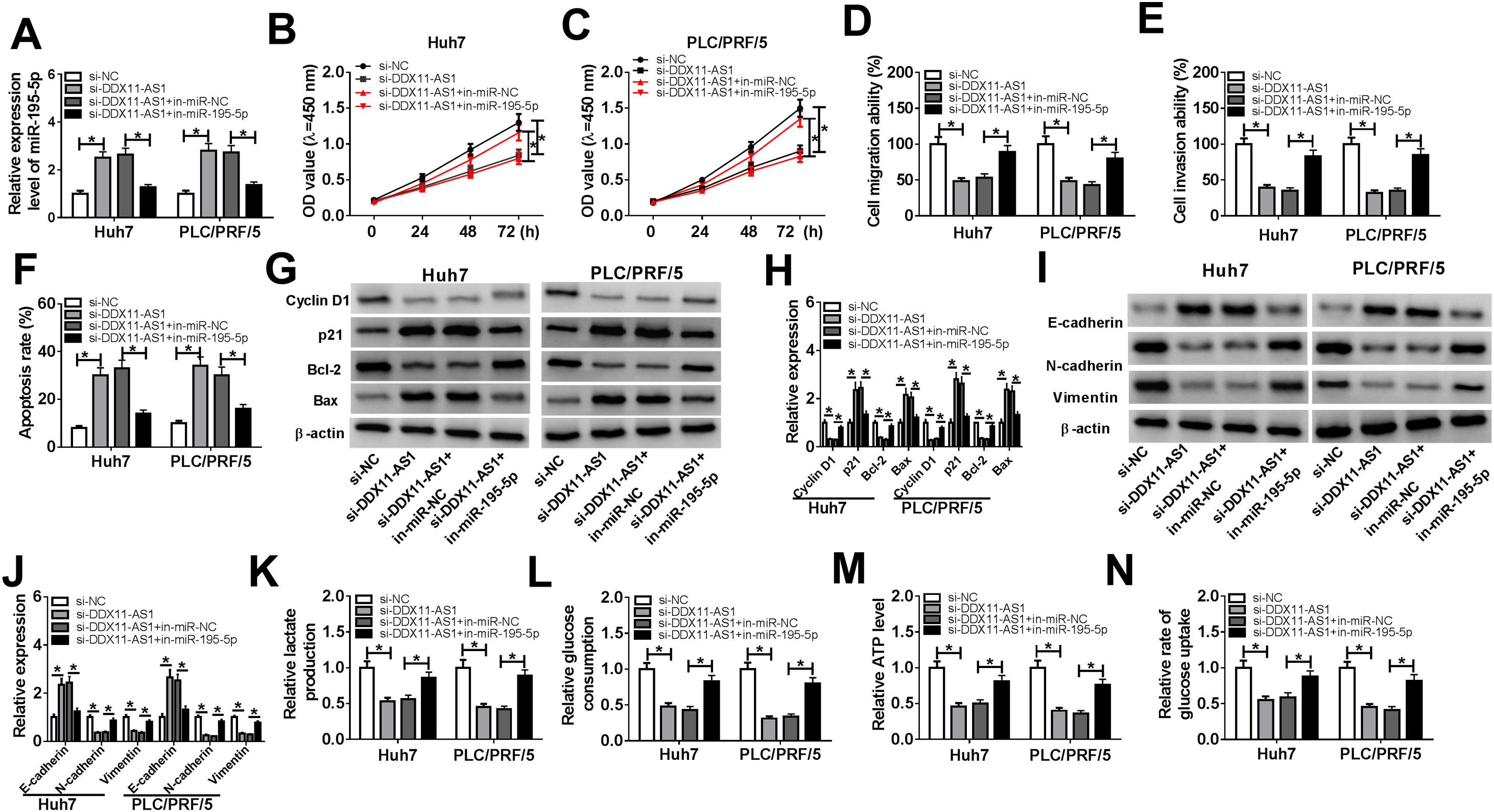
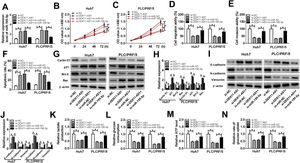
To investigate whether DDX11-AS1-mediated HCC cell progression was modulated by miR-195-5p, we co-transfected with si-DDX11-AS1 and in-miR-195-5p into Huh7 and PLC/PRF/5 cells. The increasing effect of DDX11-AS1 knockdown on miR-195-5p expression could be reversed by miR-195-5p inhibitor, which suggested that the transfections of si-DDX11-AS1 and in-miR-195-5p in HCC cells were successful (Fig. 5A). Additionally, CCK-8 and transwell assay showed that miR-195-5p silencing restored the decreasing effect of DDX11-AS1 knockdown on the proliferation, migration and invasion of HCC cells (Fig. 5B–E). Meanwhile, apoptosis determination assay results indicated that inhibition of miR-195-5p also could invert the promotion effect of DDX11-AS1 silencing on the apoptosis of HCC cells (Fig. 5F). Moreover, the inhibition effect of DDX11-AS1 knockdown on the protein levels of Cyclin D1, Bcl-2, N-cadherin and Vimentin, as well as the promotion effect on the protein levels of p21, Bax and E-cadherin also could be reversed by miR-195-5p inhibitor (Fig. 5G–J). Through measuring the lactate production, glucose consumption, ATP level and glucose uptake of HCC cells, we observed that miR-195-5p inhibitor could reverse the inhibitory function of DDX11-AS1 knockdown on the glucose metabolism of HCC cells (Fig. 5K–N). The above results proved that DDX11-AS1 regulated HCC progression by sponging miR-195-5p.
Effects of DDX11-AS1 knockdown and miR-195-5p inhibitor on HCC progression. Huh7 and PLC/PRF/5 cells were transfected with si-NC, si-DDX11-AS1, si-DDX11-AS1 + in-miR-NC, or si-DDX11-AS1 + in-miR-195-5p, respectively. (A) MiR-195-5p level in Huh7 and PLC/PRF/5 cells was measured by qRT-PCR to evaluate the transfection efficiency of si-DDX11-AS1 and in-miR-195-5p. (B–C) The proliferation of Huh7 and PLC/PRF/5 cells was evaluated by CCK-8 assay. (D–E) Transwell assay was performed to assess the migration and invasion of Huh7 and PLC/PRF/5 cells. (F) The apoptosis of Huh7 and PLC/PRF/5 cells was assessed by apoptosis determination assay. (G–J) WB analysis was used to assess the protein levels of Cyclin D1, p21, Bcl-2, Bax, E-cadherin, N-cadherin and Vimentin. The lactate production (K), glucose consumption (L), ATP level (M) and glucose uptake (N) of Huh7 and PLC/PRF/5 cells were determined by Lactate Assay Kit, Glucose Assay Kit, ATP Assay Kit and Glucose Uptake Assay Kit, respectively. *P < 0.05.
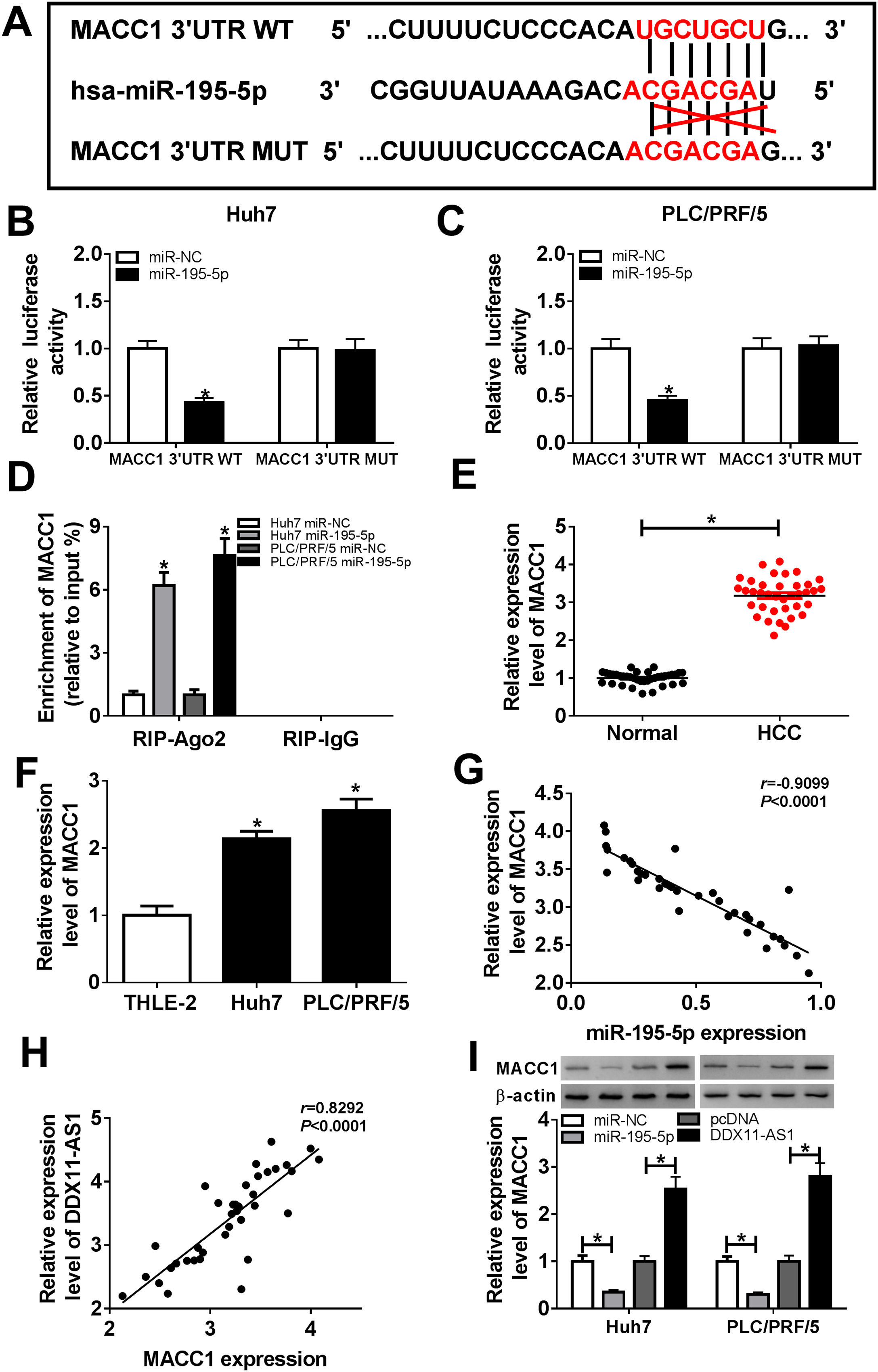
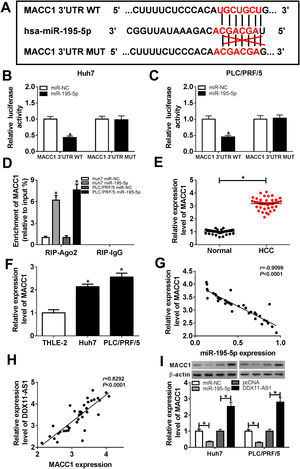
Subsequently, we conducted bioinformatics prediction (Targetscan software) to explore the downstream target genes of miR-195-5p and found that MACC1 could bind to miR-195-5p (Fig. 6A). Besides, ectopic expression of miR-195-5p suppressed the luciferase activity of MACC1 3’UTR WT reporter vector, but had no effect on that of MACC1 3’UTR MUT reporter vector in HCC cells (Fig. 6B–C). These results indicated that miR-195-5p could directly target MACC1. Furthermore, RIP analysis showed that the enrichment of MACC1 could be specifically recruited at miR-195-5p in RIP-Ago2 (Fig. 6D). Besides, the expression of MACC1 was upregulated in HCC tissues and cells detected by qRT-PCR (Fig. 6E–F). Moreover, we uncovered that MACC1 expression was negatively correlated with miR-195-5p, but negatively correlated with DDX11-AS1 in HCC tissues (Fig. 6G–H). Furthermore, miR-195-5p overexpression could inhibit the protein level of MACC1, while DDX11-AS1 overexpression could promote the protein level of MACC1 in Huh7 and PLC/PRF/5 cells (Fig. 6I).
MACC1 was a target of miR-195-5p. (A) MACC1 3’UTR WT and MACC1 3’UTR MUT containing miR-195-5p binding sites and mutant binding sites were presented. (B-C) Dual-luciferase reporter assay was used to detect the interaction between MACC1 and miR-195-5p in Huh7 and PLC/PRF/5 cells. (D) The enrichment of MACC1 in RIP-Ago2 and RIP-IgG was measured by RIP assay in Huh7 and PLC/PRF/5 cells. The expression of MACC1 was evaluated by qRT-PCR in HCC tissues (E) and cells (F). (G–H) The correlation analysis between miR-195-5p and MACC1 expression or DDX11-AS1 and MACC1 expression were determined by Pearson analysis. (I) The protein level of MACC1 was detected by WB analysis in Huh7 and PLC/PRF/5 cells. *P < 0.05.
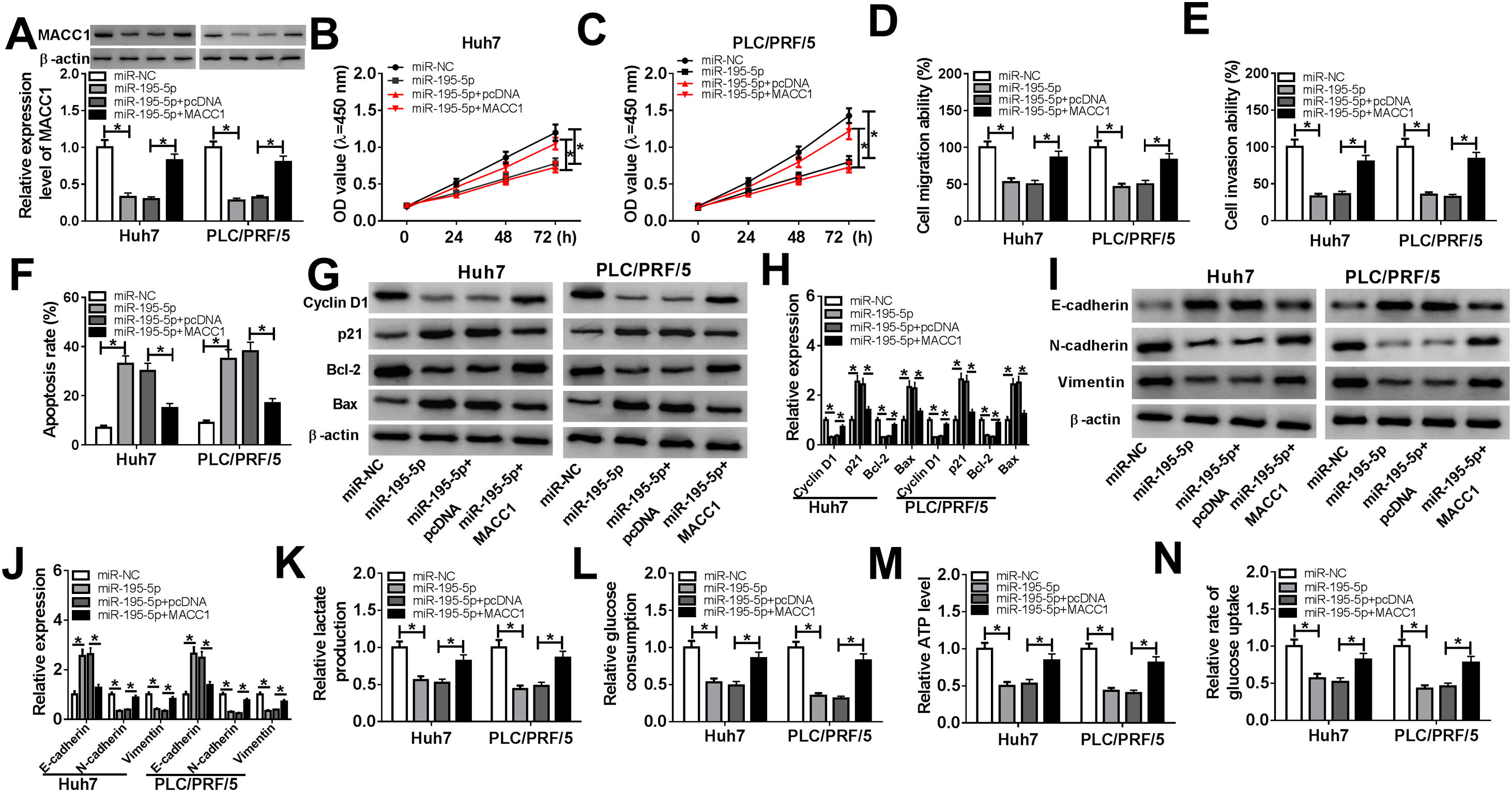
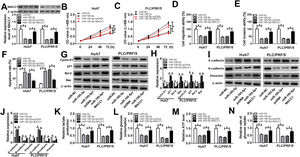
To elucidate whether miR-195-5p regulated HCC progression by targeting MACC1, we co-transfected with miR-195-5p mimic and MACC1 overexpression vector into Huh7 and PLC/PRF/5 cells. WB analysis showed that MACC1 level was blocked by miR-195-5p and promoted by MACC1 overexpression, which showed that the transfection efficiencies of miR-195-5p mimic and MACC1 overexpression vector were good (Fig. 7A). Then, we measured the biological function of HCC cells. As shown in Fig. 7B–F, the aberrant expression of MACC1 reversed the inhibition effect of miR-195-5p mimic on the proliferation, migration and invasion of Huh7 and PLC/PRF/5 cells, as well as the promotion effect of it on the apoptosis of HCC cells. Furthermore, overexpressed MACC1 also reversed the decreasing effect of miR-195-5p on the protein levels of Cyclin D1, Bcl-2, N-cadherin and Vimentin, as well as the increasing effect on the protein levels of p21, Bax and E-cadherin in Huh7 and PLC/PRF/5 cells (Fig. 7G–J). In addition, the suppression effect of miR-195-5p on the lactate production, glucose consumption, ATP level and glucose uptake of HCC cells also could be inverted by overexpressed MACC1 (Fig. 7K–N). Hence, we concluded that miR-195-5p inhibited the progression of HCC by targeting MACC1.
Effects of miR-195-5p mimic and MACC1 overexpression on HCC progression. Huh7 and PLC/PRF/5 cells were transfected with miR-NC, miR-195-5p mimic, miR-195-5p + pcDNA, or miR-195-5p + MACC1, respectively. (A) WB analysis was used to measure the protein level of MACC1 in Huh7 and PLC/PRF/5 cells to assess the transfection efficiency of miR-195-5p mimic and MACC1 overexpression vector. (B–C) CCK-8 assay was performed to assess the proliferation of Huh7 and PLC/PRF/5 cells. (D–E) Transwell assay was employed to evaluate the migration and invasion of Huh7 and PLC/PRF/5 cells. (F) Apoptosis determination assay was used to measure the apoptosis of Huh7 and PLC/PRF/5 cells. (G–J) WB analysis was performed to detect the protein levels of Cyclin D1, p21, Bcl-2, Bax, E-cadherin, N-cadherin and Vimentin. The lactate production (K), glucose consumption (L), ATP level (M) and glucose uptake (N) of Huh7 and PLC/PRF/5 cells were tested by Lactate Assay Kit, Glucose Assay Kit, ATP Assay Kit and Glucose Uptake Assay Kit, respectively. *P < 0.05.
Currently, the morbidity of HCC is on rise year by year, and the proliferation and metastasis of cancer is a major challenge for us to overcome. Therefore, it is very important for us to seek the effective therapeutic targets of cancers. The importance of cellular metabolic reprogramming in cancer has received extensive attention from scholars, especially the changes in glucose metabolism. Glucose metabolism has become an important way for cancer cells to obtain energy, so it is important to explore the mechanisms that affect glucose metabolism. In our research, we discovered that lncRNA DDX11-AS1 was increased in HCC tissues and cells, which was accorded with the conclusion of Li et al. study [21]. DDX11-AS1 silencing restrained HCC cell proliferation, invasion, migration and glucose metabolism, and promoted the apoptosis in vitro. In addition, we also demonstrated that interfering DDX11-AS1 inhibited HCC tumor growth in vivo. Therefore, our results revealed that DDX11-AS1 might function as an oncogene in HCC. In terms of mechanism, we found that DDX11-AS1 could regulate the expression of MACC1 by targeting miR-195-5p.
MiR-195-5p had abnormal expression in many cancers and was involved in the regulation of cancer, including ovarian cancer [22], lung cancer [23], and colorectal cancer [24]. Xu et al. proved that miR-195-5p suppressed cell migration, invasion and proliferation in HCC [25]. In our study, we verified that DDX11-AS1 could directly bind to miR-195-5p, and it was negatively related to miR-195-5p expression in HCC. Meanwhile, down-regulation of miR-195-5p in HCC tissues and cells was consistent with previous reports [25]. Furthermore, miR-195-5p inhibitor could recover the suppression effect of silenced DDX11-AS1 on the proliferation, migration, invasion and glucose metabolism, and the promotion effect of it on the apoptosis of HCC cells. Latest research by Huang et al. indicated that lncRNA SNHG1 could inhibit miR-195-5p expression to regulate the proliferative and migratory potentials of HCC cells [26]. This suggested that miR-195-5p had an anti-cancer effect in HCC, which was consistent with our results.
In order to decipher the mechanism of DDX11-AS1, we continued to search for the downstream targets of miR-195-5p. After prediction and confirmation, we concluded that MACC1 was the target of miR-195-5p, and it was negatively related to miR-195-5p and positively related to DDX11-AS1. MACC1 could function as an oncogene in HCC had been predicted in previous studies. Yao et al. reported that MACC1 restrained cell apoptosis and accelerated cell growth in HCC [27]. Sun et al. showed that the level of MACC1 was related to the survival rate and recurrence rate of HCC patients [28]. Moreover, knockdown of MACC1 blocked the migration and invasion of HCC cells [29]. Similarly, in this study, we revealed that overexpression of MACC1 reversed the suppression function of miR-195-5p aberrant expression on HCC cell progression, which once again confirmed that MACC1 was targeted by miR-195-5p and it participated in the regulation of miR-195-5p on HCC progression.
In summary, this study demonstrated that DDX11-AS1 promoted the progression of HCC by regulating the miR-195-5p/MACC1 pathway, which might involve in the regulation of HCC as a pro-oncogenic factor. The results of this research might provide a vital theoretical basis for HCC treatment.AbbreviationslncRNA long non-coding RNA hepatocellular carcinoma DEAD/H box protein 11 antisense RNA 1 metastasis-associated in colon cancer-1 quantitative real-time PCR cell counting kit-8 RNA immunoprecipitation western blot microRNAs
The authors declare that there is no conflict of interest regarding the publication of this article.
FundingNo funding was received.
None.