Although Entamoeba dispar displays a similar morphology to Entamoeba histolytica, cellular and molecular studies have revealed significant differences between these two amoebae, including the former being characterized as non-pathogenic and the later as pathogenic. However, recent in vivo and in vitro experiments have shown that E. dispar strains of different origin are capable of causing liver damage and destroying cell culture lines in the presence of common intestinal bacteria. These results suggested that E. dispar may present pathogenic behavior according to the specific E. dispar strain, culture and environmental conditions. To investigate this possibility, we carried out in vivo and in vitro studies using a xenic strain E. dispar (ICB-ADO) isolated from a symptomatic non-dysenteric Brazilian patient. This strain was able to induce liver necrosis in a hamster model that was more severe than that produced by E. histolytica. The ICB-ADO isolate also caused significantly more destruction of cultured MDCK cells and increased loss of transepithelial resistance than did the E. histolytica. Xenic E. dispar exhibited high proteolytic activity, which was partially inhibited by the addition of cysteine-protease inhibitors. Based on our biochemical and molecular characterization of E. dispar (ICB-ADO) xenic culture and its ability to produce liver abscesses, we conclude that this specific strain can indeed produce tissue damage, distinct from the frequently used non-pathogenic E. dispar SAW 760 strain.
Entamoeba histolytica, the causative agent of human amoebiasis, remains as an important cause of morbidity and mortality in developing countries and is the responsible for up to 100,000 deaths worldwide each year. The Entamoeba dispar species is morphologically similar to E. histolytica, but has been considered a distinct strain based on differences in the isoenzymatic patterns observed between amoebae isolates from asymptomatic patients and those with invasive disease.1 This characterization has been reinforced by several studies that have demonstrated antigenic and genetic differences between the two strains.24The presence of E. dispar in the mammalian large intestine is widely believed to constitute a parasite colonization behaving as a commensal; however, this species has been isolated from patients with symptomatic non-dysenteric colitis.5 More recently, E. dispar sequences of DNA have been detected and genotyped in samples obtained from patients with amoebic liver abscesses, suggesting that E. dispar can be also involved in the damage of human large intestine and liver.6
An important molecule that is known to be related to the pathogenicity of E. histolytica, the lectin Gal/GalNAc, has also been identified in E. dispar. However, some conformational differences exist on the E. dispar form of Gal/GalNAc lectin that may explain the reduced adherence and cytotoxicity of this strain that have been observed in vitro, and the significantly lower virulence of its trophozoites in animal models.79Interestingly, both Entamoeba species have almost the same set of genes that encode for the major virulence factors. Cysteine proteases (CP) are important virulence factors in the E. histolytica species; the production and release of these enzymes have been found to be about ten to 1,000 times higher in E. histolytica than in the E. dispar species.1011The activity of small pore-forming amphipathic peptides, known as amoebapores, in E. dispar is about one-third of that observed in E. histolytica, despite the fact that about 95% amino acid homology exists between the two strain’s amoebopores.12
Previous studies have suggested that molecular changes may be induced in E. histolytica trophozoites during the process of xenic culturing, resulting in an increased resistance to lysis by complement factors and/or modification of surface antigens of the amoeba. In this manner, the parasite may be able to effectively overcome the immune system’s ability to recognize, attack, and ultimately eliminate the parasite.1315
While it is generally accepted that the E. histolytica and E. dispar protozoans represent two distinct species, the pathogenic potential of the latter remains controversial. Recent studies have demonstrated that E. dispar trophozoites can cause intestinal focal lesions in experimental animal models and destruction of cultured monolayer epithelial cells; these findings suggest that E. dispar may induce at least some pathological changes in the human colon.16 Other recent reports have indicated that E. dispar can also produce extensive hepatic lesions in hamsters when trophozoites are administered in conjunction with the host’s original intestinal flora; the fact that these lesions are similar to those caused by E. histolytica, suggests that normal bacterial flora may interact with some E. dispar strains and influence their pathogenic behavior.51718 To date, very few experimental studies related to the use of xenic strains of E. dispar have been reported, and even fewer have focused on the role of bacteria play on influencing the pathogenic behavior of E. dispar strains.1718
In the present study, we tested the in vitro and in vivo effect of a specific E. dispar strain (ICB-ADO) that in previous preliminary studies displayed a virulence phenotype of its trophozoites in xenic culture conditions similar to the E. histolytica strain HM1-IMSS.19 Furthermore, this strain had been recently genotyped using non-coding intergenic DNA regions corresponding to tRNA genes. The four phylogenetic reconstructions performed had clearly demonstrated that the E. dispar ICB-ADO strain is a genetic variant of E. dispar species.6 We describe here our findings from the investigation into whether E. dispar presents pathogenic behavior according to the specific strains, culture and host conditions.
Material and MethodsCell cultureE. dispar trophozoites from the ICB-ADO strain (zymodeme I-non-pathogenic)5 were cultured under xenic conditions in cell culture bottles (30 mL; Corning, Lowell, MA, USA) using Pavlova’s medium formula as modified by Costa, et al.17 and containing bacterial flora. After a period of 72 h, the supernatant was removed in order to eliminate the bacterial flora and substituted with sterile PBS, pH 7.2; the mixture was incubated at 37 °C with gently shaking to wash the adhered trophozoites. This procedure was repeated once, after which the culture bottles were transferred to an ice bath for a total of 10 min. The prepared trophozoites were centrifuged at 200 x g for 5 min. The sediment amoebae were diluted in sterile PBS, pH 7.2, to obtain desired concentrations. The monoxenic strain of E. dispar trophozoiteslCB-ADO (with Crithidia fasciculata) (non-pathogenic zymodeme I) and the axenic strain of E. histolytica trophozoites HM1-IMSS (pathogenic zymodeme XIX) were cultured in TYI-S-33 medium using borosilicate glass culture tubes.20 Amoebae were harvested after 72 h of incubation by chilling the culture tubes in an ice bath for 10 min. After centrifugation at 200 x g for 5 min, the cell pellets were resuspended in fresh medium. Genetic identification of the strain was confirmed by repeating the analysis of the isoenzymatic pattern and by employing a PCR method for determining the conformational polymorphism of the fragment of 482 pb as described earlier.21
Genotyping of ADO Entamoeba isolateDNA was extracted from trophozoites (106) harvested from a xenic culture using the TRIZOL reagent kit (Invitrogen Life Technologies, Carlsbad, CA, USA) and following the manufacturer’s instructions.
In the present study the E. dispar ICB-ADO strain used had been previously re-characterized by PCR amplification of a DNA segment of the small rRNA gene subunit, consisting of 1,950 base pairs (bp) (the RD amplicon) in both E. histolytica and E. dispar.22 For amplification with RD primers, 2 μL of DNA was used as the template and 1 μL of obtained PCR product (diluted 1:10) was used as the template for amplifications with the primers Psp (specific for E. histolytica) and NPsp (specific for E. dispar).22
Both primer sets are expected to generate an 876-bp fragment that is part of the RD amplicon. The PCR reaction mix included 10 mM Tris-HCl, pH 8.3, 50 mM KCl, 0.001% gelatin, 2 mM MgCl2, 0.2 mM of each nucleotide, 0.025 U (Ampli Taq Gold, Applied Biosystems, Foster City, CA, USA) and 1 μM of the respective primer. After an initial 10 min step at 95 °C, 35 cycles consisting of 94 °C for 1 min, 55 °C for 1.5 min and 72 °C for 2 min were carried out, followed by a final step of 72 °C for 8 min.
All PCR products were electrophoresed through 1.5% agarose gels (Gibco BRL Products); the gels were then stained with ethidium bromide 0.1% solution for band visualization and photographic images were recorded.
Intrahepatic inoculation of trophozoites in hamstersMale adult golden hamsters (Mesocricetus auratus) two months-old and weighing approximately of 100 g were obtained from the Animal Center of the Center for Research and Advanced Studies of the National Polytechnic Institute, Mexico. The animals were anesthetized with sodium pentobarbital (40 mg/kg of body weight; Hypnol®, Lima, Peru) and inoculated intrahepatically with 5 × 105 trophozoites of E. dispar ADO strain (xenic culture) that had been obtained during log growth phase (72 h). All animal management protocols had been previously approved by the institutional committee. Our institution fulfills all the technical specifications for the production, care and use of laboratory animals and is certified by national law (NOM-062-Z00-1999).
Three animals were euthanized at 1, 3, 6, 12, 24, and 48 h and at the 7th day after inoculation. For controls, three individuals were inoculated with only bacteria that had been used in the xenic cultures; these animals were sacrificed at 6 and 24 h and 7 days post-inoculation. All hamsters were euthanized by an overdose of sodium pentobarbital and were handled according to the guidelines of the 2000 AVMA Panel of Euthanasia.
Liver tissue fragments were collected and fixed in 10% buffered formaldehyde, and processed for paraffin embedding. Sections were stained with hematoxylin-eosin (H&E).
Analysis of cytopathic effectMDCK monolayer cells were cultured to 80% confluence in 8-well culture chambers (Nunc Inc., Rochester, NY, USA). A minimum essential medium (MEM; Gibco-Invitrogen) supplemented with 20% fetal bovine serum (FBS; Gibco-Invitrogen) was used and the cultures incubated at 37 °C in a 5% CO2 atmosphere. Once the monolayer was formed, 3 × 105 trophozoites of E. dispar ICB-ADO strain were added and incubated for 5, 15, 30, and 60 min at 37 °C. After each time-point, cultures were washed three times with MEM without FBS, fixed with 10% methanol, stained with 5% Giemsa dye for 40 min and mounted with resin. For controls, the bacteria associated with the xenic culture of the ICB-ADO strain were used.
Microscopic images were digitalized by using a JVC TK-1270/JGB camera attached to a Nikon H500L light microscope. Morphometric analysis was performed using Kontron KS300 v.2.0 software (Kontron Electronics, Poway, CA, USA).
Transepithelial electric resistance (TER) measurementMeasurements of TER were evaluated using an EVOM epithelial voltmeter (World Precision Instruments Inc., Sarasota, FL, USA).
MDCK cells were grown in Transwell® chambers (Corning) using MEM medium supplemented with 20% FBS at 37 °C in a 5% CO2 atmosphere. The chambers were placed in a multiwell plate to form two independent compartments. After reaching confluence (approximately 100,000 cells per well), cells were washed twice with MEM without FBS and trophozoites of E. histolytica or E. dispar (at a 1:2 trophozoite:MDCK ratio) were added to the Transwell®. TER was measured at 5, 15, 30, and 60 min, and calculations were carried out based on Ohm’s law. The normalized values reported here represent the mean ± SEM at each time point (n = 6).
Protease activities in Azocoll assaysTotal crude extracts were obtained by lysing trophozoites with 5 cycles of freeze-thawing. Total protein concentration was quantified by Bradford’s method.23
Two milligrams of Azocoll (Sigma-Aldrich, St. Louis, MO, USA) were aliquoted into Eppendorf tubes (1.5 mL) containing 100 μg of crude extract; the final volume was adjusted to 500 μL with activation buffer (100 mM Tris-OH and 10 mM of CaCl2, pH 7.0). The samples were then incubated at 36 °C for 16 h. The reaction was terminated by addition of 500 μL of 10% trichloroacetic acid. Tubes were centrifuged at 4,500 × g for 5 min and supernatants were collected for spectrophotometric absorbance reading at 540 nm.
To determine the effect of inhibitors of cysteine proteases, serine proteases and metalloproteases, the following chemicals were added directly to the extracts and the mixtures allowed to incubate at room temperature for 1 h prior to addition of the substrates: p-hydroxy-mercuribenzoic acid (PHMB; 10 mM), phenylmethylsuphonyl fluoride (PMSF; 5 mM) or ethylenediaminetetracetic acid (EDTA; 2 mM), respectively. The experiments were performed in triplicate for each sample and statistical analysis was carried out using the Bonferroni test to identify significant differences among protease activities.
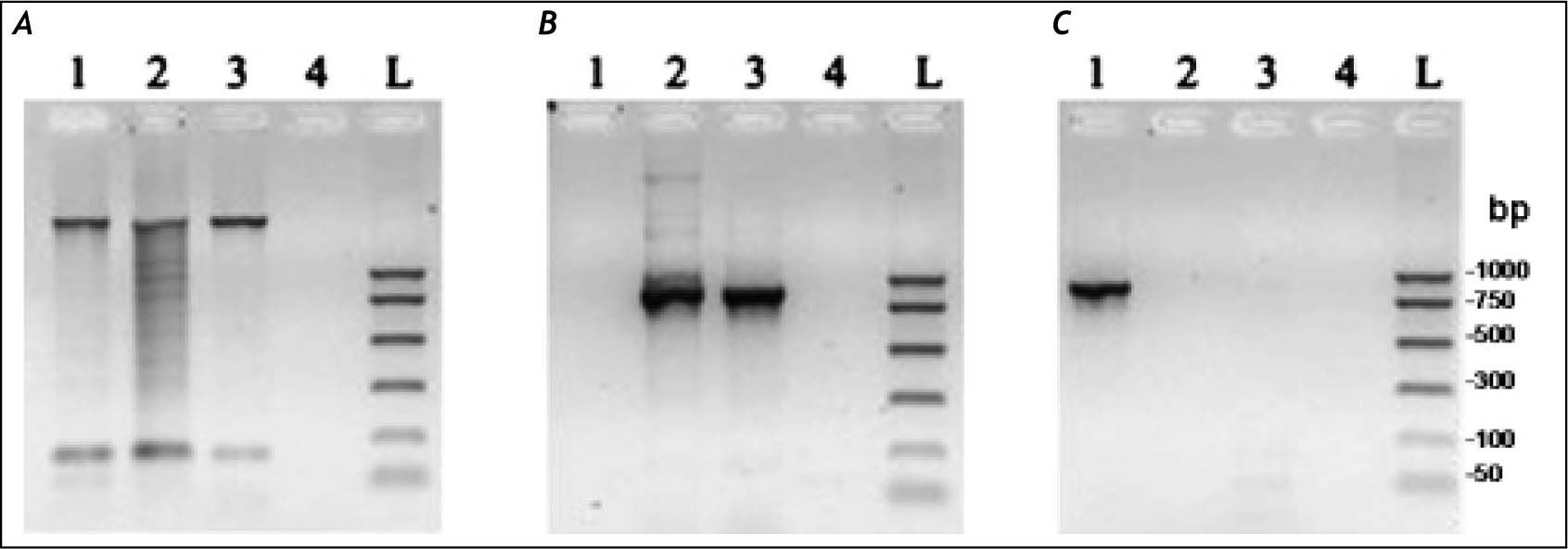
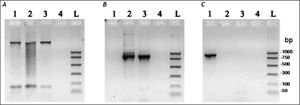
ResultsMolecular characterization of the ADO isolateThis isolate had been obtained from the feces of a Brazilian symptomatic patient clinically diagnosed with non-dysenteric colitis. Cysts from the same patient were cultured in Pavlova’s medium modified by Costa, et al., as previously described,17 and cultures were maintained in xenic conditions. The major bacterial components of this culture were determined to be Escherichia coli, Acinetobacter lwoffii and a rough E. coli colony that were presumed as components of the patient’s original intestinal flora. Previously, this isolate was genetically identified by Tannich, et al.3 as an E. dispar species, based on the conformational polymorphism of the 482 bp DNA fragment of the m17 gene that was amplified with the primers P1-S17 (forward) and P1AS20 (reverse). In our study we sought to re-test the Entamoeba species of the ICB-ADO isolate by targeting the gene for the small subunit of rRNA, as described in table 1. Figure 1A shows the PCR products that were obtained with the RD primer which was expected to amplify a 1950 bp segment of DNA in both E. histolytica and E. dispar species.22 Our amplification products agreed with the previous genotyping which identified the ICB-ADO strain as an E. dispar genetic variant.6
Primers used for genotyping.
| Primer name | Gene | Sequence (5’ to 3’) | Size range (bp) | Conditions |
|---|---|---|---|---|
| RD5 | SS rRNA | GGAAGCTTATCTGGTTGATCCTGCCAGTA | 1950 | 94 °C 1.0’ |
| RD3 | E.h./E.d. | GGGATCCTGATCCTTCCGCAGGTTCACCTAC | 55 °C 1.5’ | |
| 72 °C 2.0’ | ||||
| Psp5 | SS rRNA | GGCCAATTCATTCAATGAATTGAG | 876 | 94 °C 1.0’ |
| Psp3 | E.h. | GGGATCCTGATCCTTCCGCAGGTTCACCTAC | 55 °C 1.5’ | |
| 72 °C 2.0’ | ||||
| NPsp5 | SS rRNA | GGCCAATTTATGTAAGTAAATTGAG | 876 | 94 °C 1.0’ |
| NPsp3 | E.d. | CTTGGATTTAGAAACAATGTTTCTTC | 55 °C 1.5’ | |
| 72 °C 2.0’ |
PCR amplification of the xenic strain ICBADO and reference strains of species E. dispar SAW760 and E. histolytica HM1:IMSS. Primer RD was used to amplify DNA of species from the genus Entamoeba (A). Primer NPsp was specific for E. dispar species (B) and Psp primer was specific for E. histolytica species (C). Line 1: DNA from E. histolytica HM1:IMSS strain; line 2: DNA from E. dispar SAW760 strain; line 3: DNA from xenic ICB-ADO strain; line 4: negative controls without DNA; line L: DNA size markers.
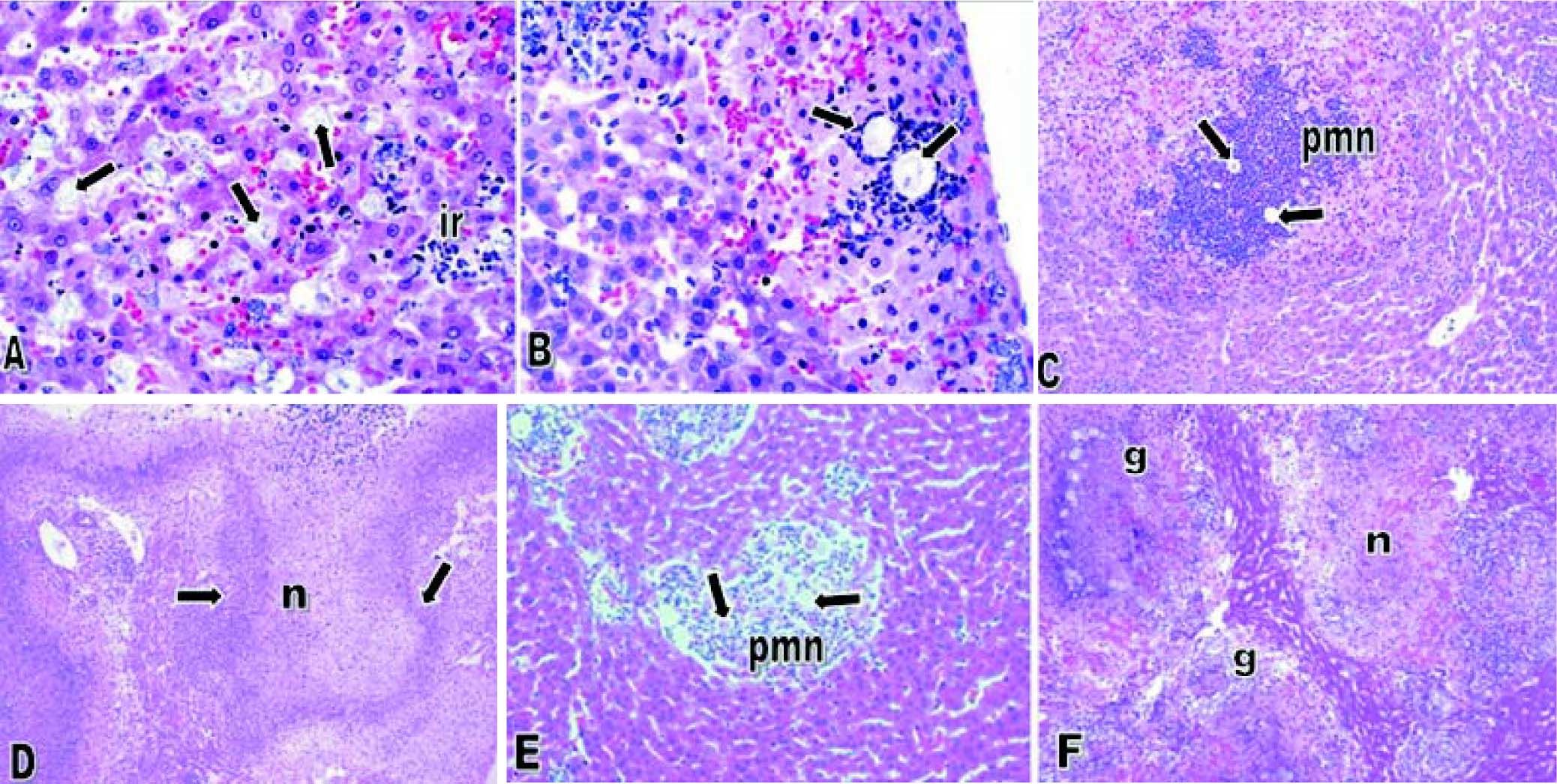
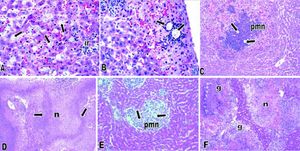
E. dispar xenic strain ICB-ADO was found to cause extensive lesions similar to those reported by others for the strain E. histolytica HM1-IMSS at seven days post-inoculation.1719 However, during the initial periods of infection the speed of abscess development observed was slower. Histological analysis indicated that liver lesions produced by ICB-ADO trophozoites at 3 h showed an important number of parasites in close contact with hepatocytes. Moreover, many of these parenchymal cells presented visual signs of cell damage, as shown in figure 2A. At 6 h post-inoculation, the amoebae induced a discrete inflammatory reaction, and the liver parenchyma proximal to the Glisson capsule appeared damaged (Figure 2B). At 12 h post-infection, the necrotic areas increased in size and numbers of inflammatory foci surrounding the amoebae were also more severe (Figure 2C). At 48 h, extensive areas of liver necrosis with scarce number of trophozoites were seen (Figure 2D).
Amoebic liver abscesses produced by E. dispar ICB-ADO (A-D) and E. histolytica HM1-IMSS (E-F). A. Three hours postinoculation of E. dispar trophozoites. Numerous amoebae were (arrows) in close contact with hepatocytes and scarce inflammatory reaction (ir) was observed. Magnification: 60 ×. B. Six hours post-E. dispar inoculation. Trophozoites (arrow) are surrounded by a discrete inflammatory infiltrate; associated hepatocytes show signs of damage. Magnification: 60 ×. C. At 12 h post-inoculation of E. dispar trophozoites, liver parenchyma showed extensive necrotic areas and important number of inflammatory cells (PMN) associated with amoebae (arrows). Magnification: 20 E. D. Forty-eight hours post-E. dispar inoculation. The areas of necrosis (n) increased significantly with chronic inflammatory infiltrates (arrows) bordering the damaged parenchyma. Magnification: 10 ×. E. Hepatic lesions were produced by E. histolytica trophozoites at 12 h post-inoculation. Several trophozoites (arrows) were surrounded by polymorphonuclear leukocytes (PMN) with some hepatocytes showing signs of damage. Magnification: 40 ×. F. At 48 h post-inoculation with E. histolytica typical granulomatous reaction (g) was observed and associated with significant necrotic areas (n). Magnification: 10 ×. All tissues were stained with H&E.
When trophozoites of the E. histolytica HM1: IMSS strain was intrahepatically inoculated, several trophozoites were found to be surrounded by inflammation at 12 h; although, the tissue damage was less extensive than with the ICB-ADO strain (Figure 2E). A granulomatous reaction was more clearly observed with the E. histolytica liver lesions than with the E. dispar ICB-ADO strain at 48 h post-infections (Figure 2F). The strain ICB-ADO that had been monoxenically cultured was unable to cause lesions in the hamster liver (data not shown).
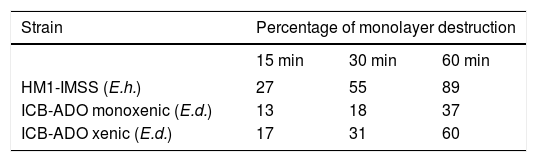
Cytophatic effect of E. histolytica and E. dispar strains on MDCK cellsE. dispar monoxenic strain ICB-ADO produced a smaller extent of damage to the MDCK monolayer cells (< 50%) as compared with that of E. histolytica strain HM1: IMSS (Table 2). E. dispar xenic strain ICB-ADO produced more cell damage than its corresponding monoxenic strain. However, this damage was less extensive than that produced by E. histolytica strain HM1-IMSS.
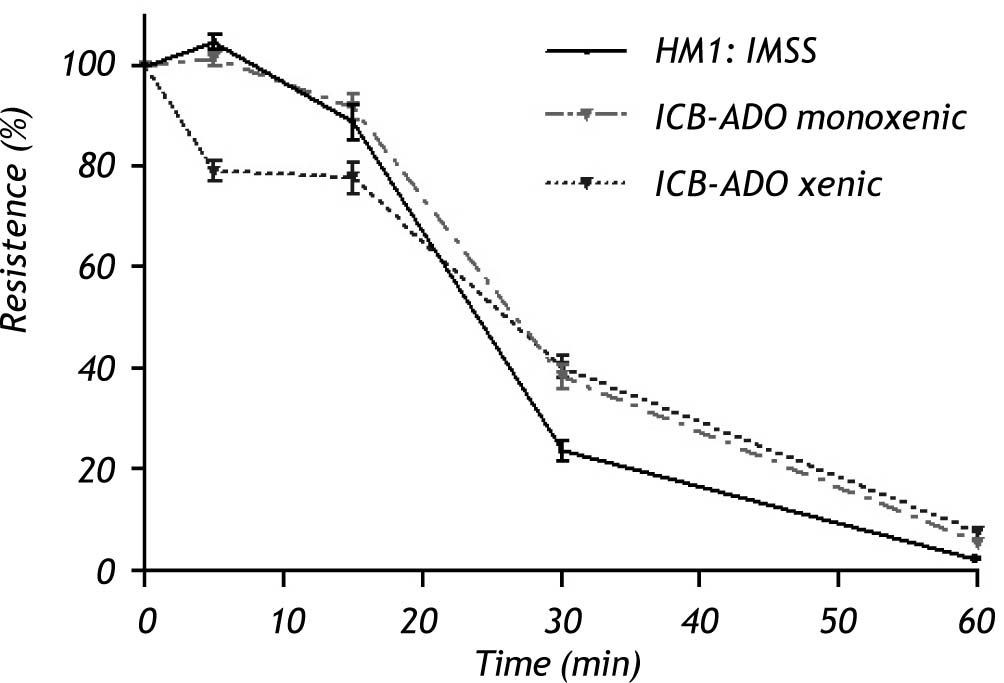
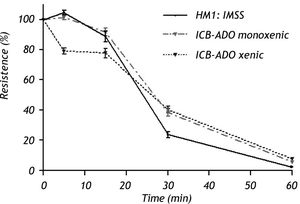
Electrophysiological measurementsThe decrease in TER observed when using xenic E. dispar ICB-ADO strain trophozoites was slower than with the E. histolytica strain. At 30 min, measurements of TER on E. dispar monoxenic and xenic ICB-ADO strains were larger than that observed for E. histolytica strain HM1:IMSS. However, after 60 min of interaction all the tested strains presented a total loss of TER (Figure 3).
Effect of E. dispar (monoxenic and xenic ICB-ADO strains) and E. histolytica (axenic HM1: IMSS strain) trophozoites on the transepithelial electrical resistance of MDCK cell monolayers. Trophozoites of each species were added at a 1:2 trophozoite:cells ratio. A faster decrease of TER was observed in E. histolytica when compared with E. dispar. At 60 min, all strains showed a loss in TER.
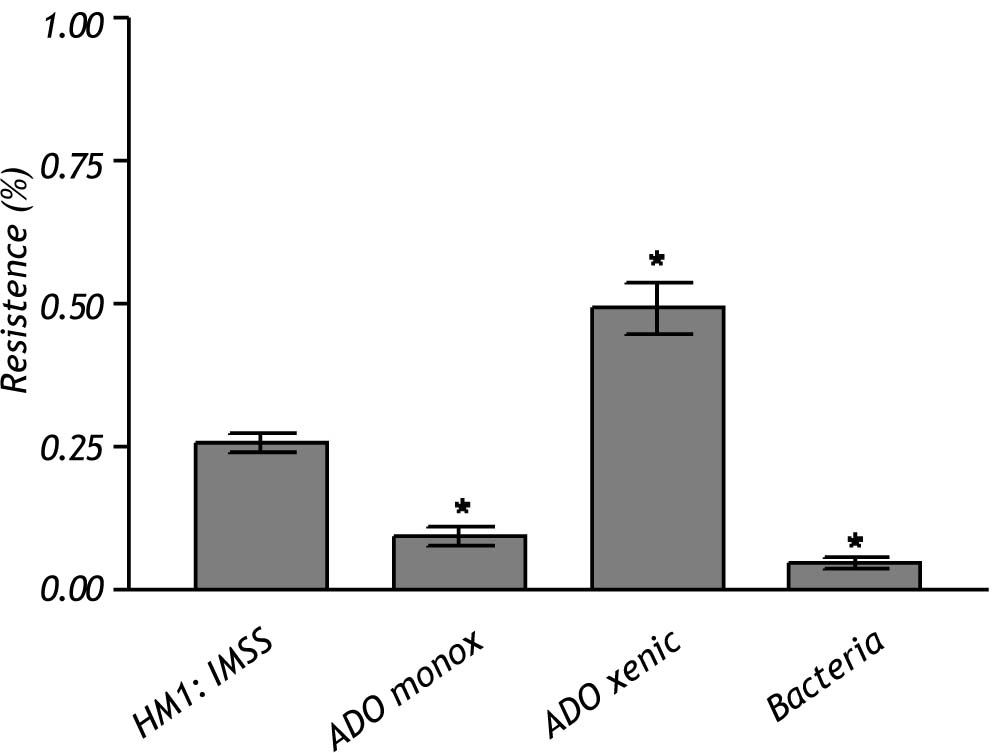
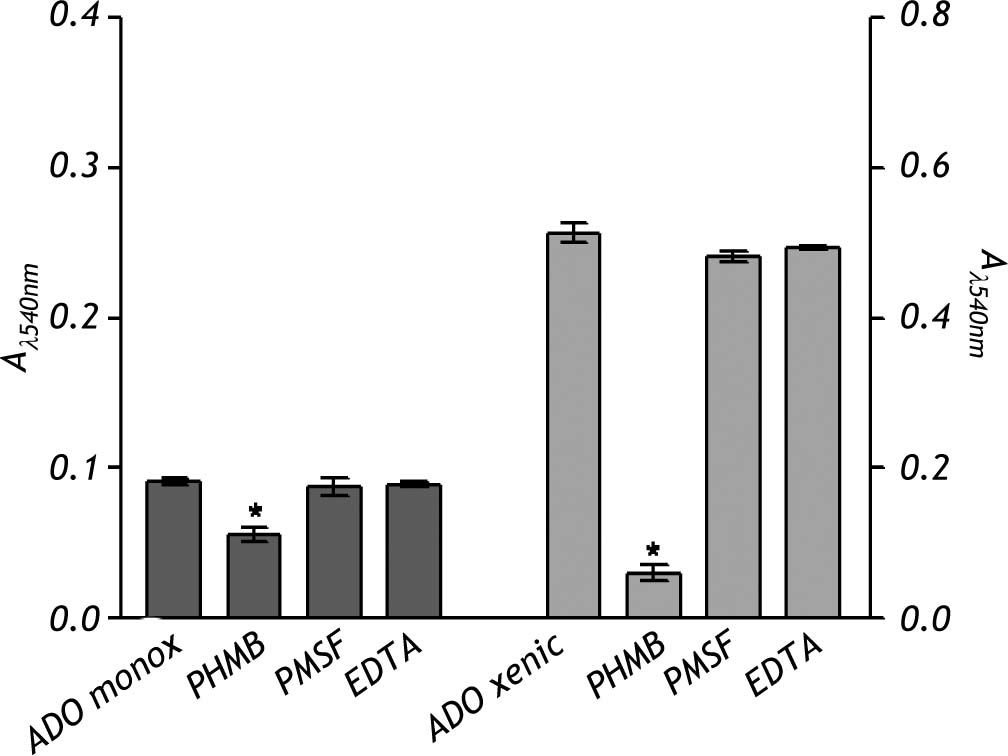
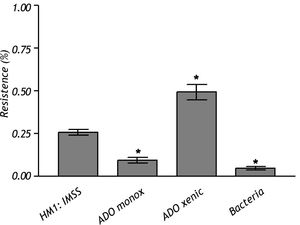
The crude extract of monoxenic E. dispar ICB-ADO strain showed lower proteolytic activity than the E. histolytica strain HM1: IMSS. However, the crude extract from the xenic ICB-ADO culture exhibited stronger proteolytic activity than the E. histolytica strain (Figure 4).
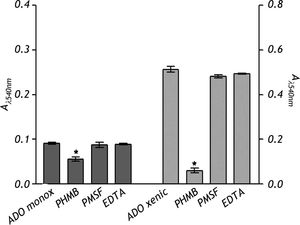
The nature of the proteolytic activity observed in the crude extracts was determined by using specific inhibitors for cysteine, serine and metalloproteases. The substrate used for all the inhibitors was the nonspecific protease azo-dye-impregnated collagen (Azocoll). The inhibitor for cysteine protease PHMB had a significant inhibition capacity over the pro-teolytic activity of the xenic strain (Figure 5). The same occurred in the case of the monoxenic strain, although with a less intensity. The PMSF and EDTA inhibitors of serine and metalloproteases did not reduce the proteolytic activities.
DiscussionMost of the current knowledge regarding the biology and physiopathology of E. dispar is based on a unique species isolated from the feces of a single English male who was an asymptomatic cyst passer; this strain is classified as SAW760 and cultured axenically.24 However, little is known about other isolates of the genetically similar E. dispar species that has been cultured monoxenically or xenically by other groups.5,16,1725 Previous work on the pathogenicity and virulence of E. dispar support the idea that this strain, in certain conditions, such as alterations of the immune system or interactions with bacteria of host intestinal flora, can produce a pathogenic behavior in humans, contrarily as was originally proposed by Brumpt26 that E. dispar was always a non-pathogenic strain. Moreover, there are some reports in the literature using other different isolates of E. dispar, such as those described by Costa, et al.,17 Furst, et al.18 and Costa, et al.,25 which have shown the production of liver damage in experimental animals. The scarce information available for the biological behavior of other strains, different from the original E. dispar SAW760, has been a direct result of the technical difficulties associated with axenic culture conditions, and has impeded our understanding of the pathogenic capabilities of this species. Thus, it is necessary to perform more experimental studies using several E. dispar isolates to attain a better understanding of their pathogenic potential and to determine whether this species is capable of invading the mammalian intestine and liver.
To this end, we investigated the E. dispar ICB-ADO strain that was originally isolated from a stool sample of a symptomatic Brazilian patient with non-dysenteric colitis. This particular isolate has been successfully cultured by others in monoxenic and xenic conditions17 and has been genotyped using different targets of polymorphic intergenic DNA region corresponding to tRNA genes (SQD3-5, Dsp5-6, StgaD35, and NKD3-5). The phylogenetic reconstruction performed by the Neighbor-Joining method for E. dispar, E. histolytica and ICB-ADO strains using the sequences of PCR products obtained with the appropriated primers clearly showed that the ICB-ADO strain is included in the E. dispar group in all phylogenies.6 In addition, this work demonstrates the presence of genetic markers of E. dispar in aspirates from liver abscess of different patients, indicating the possible invasive role of E. dispar species. The authors discuss the importance of a new data observed in E. dispar emphasizing the importance of this species in the damage of intestine and liver. Therefore, our results introduce again the important question: whether E. dispar is a simple commensal parasite (Brumpt hypothesis) or is capable to produce, in certain conditions, important lesions as described by several authors? We have raised four important hypotheses regarding the physiopathology of the amoebic liver abscess:
- •
First, E. histolytica invades the intestinal mucosa and then, both species (E. histolytica and E. dispar) reach the liver through the portal circulation. E. dispar does not produce tissue damage but may take advantage of the pathogenic capacity of the E. histolytica species.
- •
Second possible explanation proposes that the two species are similarly responsible of both the intestinal and liver damage.
- •
Third hypothesis deals with bacteria-mediated pathogenicity. This suggests that the pathogenesis of at least some E. dispar strains may be mediated by a type of bacterial flora in a particular host flora.
- •
The last hypothesis suggests that a process of recombination event between E. histolytica and E. dispar species may be occurring.
Therefore, these previously mentioned hypotheses are introducing again to the highly important discussion, whether E. dispar is just a commensal strain in the human gut or it has the ability to invade tissues and organs.
The induction of amoebic liver abscess (ALA) in the hamster model (susceptible to liver damage) was performed by injecting the xenic trophozoites of this ICB-ADO strain and resulted in serious hepatic damage, surpassing 40% of the total organ weight by 7 days post-infection. This unexpected result led us to question the variable pathogenic potential of all available strains of E. dispar. The previously reported histological events preceding the formation of the liver abscess in hamsters27-29 served as a basis by which we compared the morphological and other biological findings after inoculation with the E. dispar ICB-ADO strain. In the early stages of infection (1-12 h) we observed inflammatory foci that gradually increased in size and number; a small number of amoebae were associated with the inflammation and some showed an atypical morphology. One of the major findings at 6 h after intrahepatic inoculation was the presence of trophozoites in close contact with hepatocytes where the liver parenchyma displayed signs of damage (unpublished results). These findings differed from those published by Tsutsumi, et al.26 and Chadee & Meerovitch,30 which indicated that amoebae surrounded by inflammatory infiltrates were lysed and in turn damaged the hepatic parenchyma. Another important difference with the previous reports of ALA produced by E. histolytica in hamster and gerbil models2830 is that in our E. dispar infected hamsters the granulomatous reaction was practically absent. The liver lesions produced in our study by the E. dispar ICB-ADO strain (although with slow evolution at the beginning) progressed in a rapid manner after 24 h post-intrahepatic inoculation and resulted in extensive necrotic areas, as compared to the damage induced by E. histolytica HM1: IMSS.
The possible role of host associated bacteria present in ICB-ADO strain cultured xenically was discarded after inoculating only the bacterial flora in hamster livers. This procedure resulted only in a small periportal inflammatory infiltrate in animals euthanized at 7 days post-inoculation. In addition, inoculation of E. dispar monoxenic strain ICB-ADO did not cause any damage to the liver of the animals, only a scarce inflammatory reaction was observed in the portal areas (data not shown).
Studies reported by Espinosa-Cantellano, et al.16 using an axenic strain of E. dispar (strain SAW) demonstrated that trophozoites of this specific strain presented a discreet cytopathic effect on MDCK cells, requiring double the time to reach equal E. histolytica capacity to completely break down the transepithelial electrical resistance. To confirm the pathogenic behavior of our E. dispar species (ICB-ADO), we used MDCK cells and estimated the cell damage by measuring the TER. This technique is a highly sensitive procedure to detect the early damages produced by trophozoites in a well-established and controlled cell monolayer.31 The extent of cell damage depends on the strain and the trophozoite/epithelial cells ratio. We observed that E. dispar ADO xenic strain presented less damage to MDCK cells than did the E. histolytica strain HM1-IMSS.
Some data have suggested that adhesion constitutes an important and crucial phenomenon during the early cell damage by E. histolytica trophozoites.3233 It is possible that in the case of E. dispar in xenic culture, at the beginning of an in vitro interaction with MDKC cells, the amoeba-bacteria association may act to physically interfere with the adhesion capability to cell monolayers. A similar work carried out by Costa, et al.,17 using VERO cell line cultures, indicated a small degree of cell destruction occurred. However, at later stages the presence of bacteria associated with amoeba may instead stimulate some virulence factors produced and secreted by the parasite, such as the proteases; this phenomenon may characterize the pathophysiology of intestinal amoebiasis.34
To examine the proteolytic activity of this amoeba as mechanisms involved in tissue destruction, we used Azocoll as a chromogen substrate. The E. dispar ICB-ADO xenic strain displayed a strong proteolytic activity, a particularly surprising result since E. dispar is widely considered to be a poor protease producer.35 Since the bacteria alone were unable to produce significant protease activity, the protease activity observed in the ADO-ICB xenic strain was presumed to be due to the trophozoites of E. dispar.
Several studies have suggested that bacteria/E. histolytica association has a significant effect on the trophozoites behavior, thus modulating the phenotypical changes and virulence properties of amoebae.13,33,3638 Studies reported by Bruchhaus, et al.39 demonstrated that only three (ehcp1, ehcp2, ehcp5) of 50 genes of cysteine proteases identified in the E. histolytica genome40 were strongly expressed under culture conditions. However, Singh, et al.41 demonstrated that the association of E. histolytica/ E. coli, resulted in a strong activity of cysteine proteases, a high expression of the ehcp2 and ehcp5 genes, and significant cellular cytotoxicity. The regulation of these genes in the proteolytic pathway may also occur in some strains of E. dispar when associated with bacteria.
Early studies conducted through the interaction of E. histolytica trophozoites with bacteria have shown that after this interaction the virulence of the amoebae was amplified, increasing their cytopathic effect to epithelial cells and amoebic liver abscesses in hamster model.4244 Recent studies by Galván-Moroyoqui, et al.34 have demonstrated that following association of E. histolytica with enteropathogenic bacteria there is an increase of Gal/Gal-NAc lectin expression on the surface of trophozoites, which results in an increased adhesion capacity and higher cytopathic effect. Moreover, these authors demonstrated that the presence of pathogenic entero-bacteria could induce changes in epithelial cells (production of pro-inflammatory cytokines) that facilitate adhesion capacity and tissue damage induced by the invasive trophozoites.
It has been proposed that bacteria in the gastrointestinal tract may be one of the factors that modify or modulate the virulence of the trophozoites of E. histolytica, experiments in vitro have shown that pathogenic bacteria can increase the virulence of E. histolytica causing further damage and adhesion to target cells.34 In this context it is possible that E. dispar naturally interact with pathogenic bacteria in the intestinal tract, however, previous reports have shown that the interaction of E. dispar SAW760 with pathogenic bacteria does not change the virulence of this strain of amoeba.34 In contrast, here we show that xenical culture with bacteria increases the virulence of E. dispar ADO causing damage to cultured epithelial cells (MDCK), enhancing the protease activity and producing larger amoebic liver abscess in hamsters.
Studies using the xenic strains of E. dispar are scarce in the literature1718, 25 and, of those, very few have sought to correlate or explain the influence of bacteria on these E. dispar strains. The emerging recognition of the functional relationships that exist between commensal communities, the host immune system, and the susceptibility to inflammatory diseases emphasizes the need for a better understanding of the pathways that initiate and maintain amoebae-bacterial symbiosis.
The paradigm that only E. histolytica should be considered as a potential pathogen that can cause serious damage to the large intestine and extraintestinal organs must be reconsidered, since recent studies have shown that E. dispar can act as a potential agent capable to produce tissue damage in humans. In the work presented herein we have demonstrated that E. dispar may assume a pathogenic capacity when cultured in xenic conditions. This can take place during the colonization of the gut by this amoeba, which interacts not only with commensal bacteria but also with pathogenic bacteria, which could have an important role in the invasive form of the disease. Our results reinforce the existence of potentially pathogenic strains of E. dispar, and this in turn may change the epidemiological situation (based on molecular biology findings) of the complex E. histolytica/E. dispar with an important impact in the treatment, similarly as occurs with patients with E. histolytica infection. Currently, we are carrying out additional studies focused on the specific role of bacteria related to the pathogenesis of E. dispar. In summary, our results strongly suggest that the presence of bacteria could contribute to the regulation of virulence factors, including protease activities and, consequently, may have an important impact on the manifestation of virulence by E. dispar ADO strain xenic culture.
Abbreviations- •
Gal/GalNAc: Galactose/N-acetyl Galactosamine.
- •
PBS: phosphate buffer saline.
- •
MDCK: Madin-Darby canine kidney.
- •
MEM: minimum essential medium.
- •
FBS: fetal bovine serum.
- •
TER: transepithelial resistant.
- •
PHMB: P-Hydroxy-MercuriBenzoic acid.
- •
PMSF: PhenylMethylSuphonyl fluoride.
- •
EDTA: EthyleneDiamineTetracetic acid.
We wish to thank to Joáo da Costa Viana, Edna Maria Pires, Silvia Galindo-Gómez, Liliana Rojas and Patricia Morán for their valuable technical assistance.
Financial SupportThis work was partially support by Grant No 79220 from the National Council for Sciences and Technology (CONACyT) Mexico and Foundation for Research Development of Minas Gerais State (FAPEMIG), Brazilian Coordination for the Improvement of Higher Education Personnel (CAPES).