This study aims to compare reported proteins expressed in controls and patients with SS in tear, saliva and serum from proteins identified in papers published between 2000 and 2020, to obtain a systematic comparison of proteins expressed in SS patients to find proteins that can be used as biomarkers in SS. Differentially expressed proteins from Sjögren's patients identified in the studies were classified by body fluid (saliva, serum, tear and saliva from healthy patients). Protein pairing was carried out in RStudio, using the previously created data framework, PANTHER and REACTOME 7.O. were used to explore the type of proteins associated with SS and enriched metabolic pathways. Five differentially expressed proteins were found in saliva, tear, and serum. Finally, this study suggests antithrombin III and E-cadherin. as biomarkers in SS, because these proteins are differentially expressed in plasma, blood, and tear, and that neutrophil degranulation pathway as a target for SS.
Este estudio tiene como objetivo comparar las proteínas reportadas como diferencialmente expresadas en pacientes con síndrome de Sjögren (SS), en lágrima, saliva y suero, versus controles, a partir de proteínas identificadas en artículos publicados entre 2000 y 2020, a fin de encontrar una comparación sistemática de la proteína expresada en pacientes con SS y poder hallar proteínas que se puedan utilizar como biomarcadores en SS. Las proteínas expresadas diferencialmente de los pacientes de Sjögren identificadas en los estudios se clasificaron por fluido corporal (saliva, suero, lágrimas y saliva de pacientes sanos). El emparejamiento de proteínas se realizó en RStudio, utilizando el marco de datos previamente creado, y se utilizó Panther y Reactome 7.O. para explorar el tipo de proteínas asociadas con SS y las vías metabólicas enriquecidas. Se encontraron 5 proteínas expresadas diferencialmente en saliva, lágrimas y suero. Este estudio muestra que la antitrombina iii y la E-cadherina son biomarcadores en SS, como también que la vía de desgranulación de neutrófilos es la diana para SS.
Sjögren's syndrome (SS) is a systematic autoimmune disease characterized by a lymphoid infiltrate in the exocrine glands, that affects mainly in tear and salivary glands causing a decrease in the secretion, which are known as xerophthalmia and xerostomia.1 This disease can be classified as primary or secondary syndrome (Polyautoimmunity), the first one being an isolated pathology and the later when it is associated to other diseases such as rheumatoid arthritis, systemic lupus erythematosus, inflammatory myopathy and systemic.2
The etiology of Sjögren's syndrome is complex, it is characterized by a lymphocytic infiltration of the exocrine glands and an exaggerated reaction of the activated T and B lymphocytes. The epithelial cells suffer apoptosis induced by signals from T cells. Infiltrated lymphocytes not only induce apoptosis on epithelial cells, they also increase production of proinflammatory cytokines to extend the autoimmune injury.3
Diagnosis of Sjögren's syndrome used to be realized according to six criteria proposed in 2002, known as the AECG-criteria (American-European Consensus Group), which were: oral and eye symptoms related to dry mucous membranes, eye signs to objectively evaluate the integrity of the ocular conjunctive, the histopathology defined by the presence of inflammatory cell infiltration, involvement of salivary glands determined by secretion of saliva, sialography or scintigraphy and the presence of antibodies anti-Ro or anti-La.4 ACR criteria to define the SS when the weights from the 5 criteria items below are summed: Labial salivary gland with focal lymphocytic, anti-SSA/Ro positive, ocular Staining Score ≥5, Schirmer's test ≤5mm/5min in at least 1 eyes and unstimulated whole saliva flow.5
Diagnosis of this disease is complex due to the lack of a differential test, also an invasive procedure is needed in order to construct a diagnostic approach, making diagnostic process fearsome for patients. Because of this, with the advances in genomics and proteomics, the aim is to find specific biomarkers for the diagnosis to be sensitive enough to predict the disease's development.5
Currently, genomic and proteomic studies exist, which have identified proteins expressed in patients with SS. Kuo et al. carried out a case-control study in which they found a major concentration of MMP-9 and LTF (Lactotransferrin, associated with eye inflammation), but also a decrease of Lidocaine 1 and Lactrine, the later related to the increase of both MMP-9 (matrix metallopeptidase 9) and LTF.6
Extracellular proteins from tear and saliva vesicles of SS patients were analyzed, and an overexpression of proteins associated with lymphocyte mediated immunity, neutrophil signaling, calcium receptors and in particular the highlight expression of lipocaline-associated neutrophil gelatinase.7
Various studies demonstrate that patients with dry eye and SS have an altered proteomic profile that involve processes such as immunity, inflammation and oxidative stress.8 This studies also show that those alterations can be identified in different fluids like saliva, serum, tear and CSF (cerebrospinal liquid).9–11
There is currently an approach to integrate reported protein data to explore biological events in a holistic way.12 In a tentative approach to this methodology, this study aims to compare reported proteins expressed in controls and patients the with SS in tear, saliva and serum from proteins identified in papers published between 2000 and 2020, to obtain a systematic analyze of proteins expressed in SS patients with the purpose to find proteins that can be used as biomarkers in SS (Fig. 1).
Material and methodsSearch for data sets of identified proteins in tears, saliva and serum of Sjögren patients was done using PubMed's database (www.nlm.nih.gov/bsd/pubmed.html). Key words used were Sjögren and/or saliva, serum and tear. Studies from 2000 to 2020 were selected, excluding studies that reported animal or bacterial data; we also selected studies with a LC-MS (Liquid chromatography–mass-spectrometry) analysis.
Data analysisDifferentially expressed proteins from Sjögren patients identified in the studies were classified by body fluid (saliva, serum, tear and saliva from healthy patients) (Table 1). The same process was done for proteins referenced in each article.
Before any analysis was done, a database was created in which every list of proteins, of every article was identified. The database included protein ID, protein name, fluid type and the group that it belonged to (patient or control). Redundant IDs or unidentified sequences were eliminated.
Protein pairingProtein pairing was carried out in RStudio, using the data frame previously created, in order to identify proteins expressed between Sjögren patients, and proteins expressed in control groups that were also expressed in Sjögren patients.
Data and protein class visualizationPANTHER (Protein ANalysis THrough Evolutionary Relationships http://www.pantherdb.org/) and REACTOME 7.O. (https://reactome.org/PathwayBrowser/#TOOL=AT) were used in order to explore the type of proteins associated with SS and predominantly enriched metabolic pathways, as a result it is obtained an overrepresentation analysis: a statistical (hypergeometric distribution) test that determines whether certain pathways are over-represented (enriched) in the submitted data.
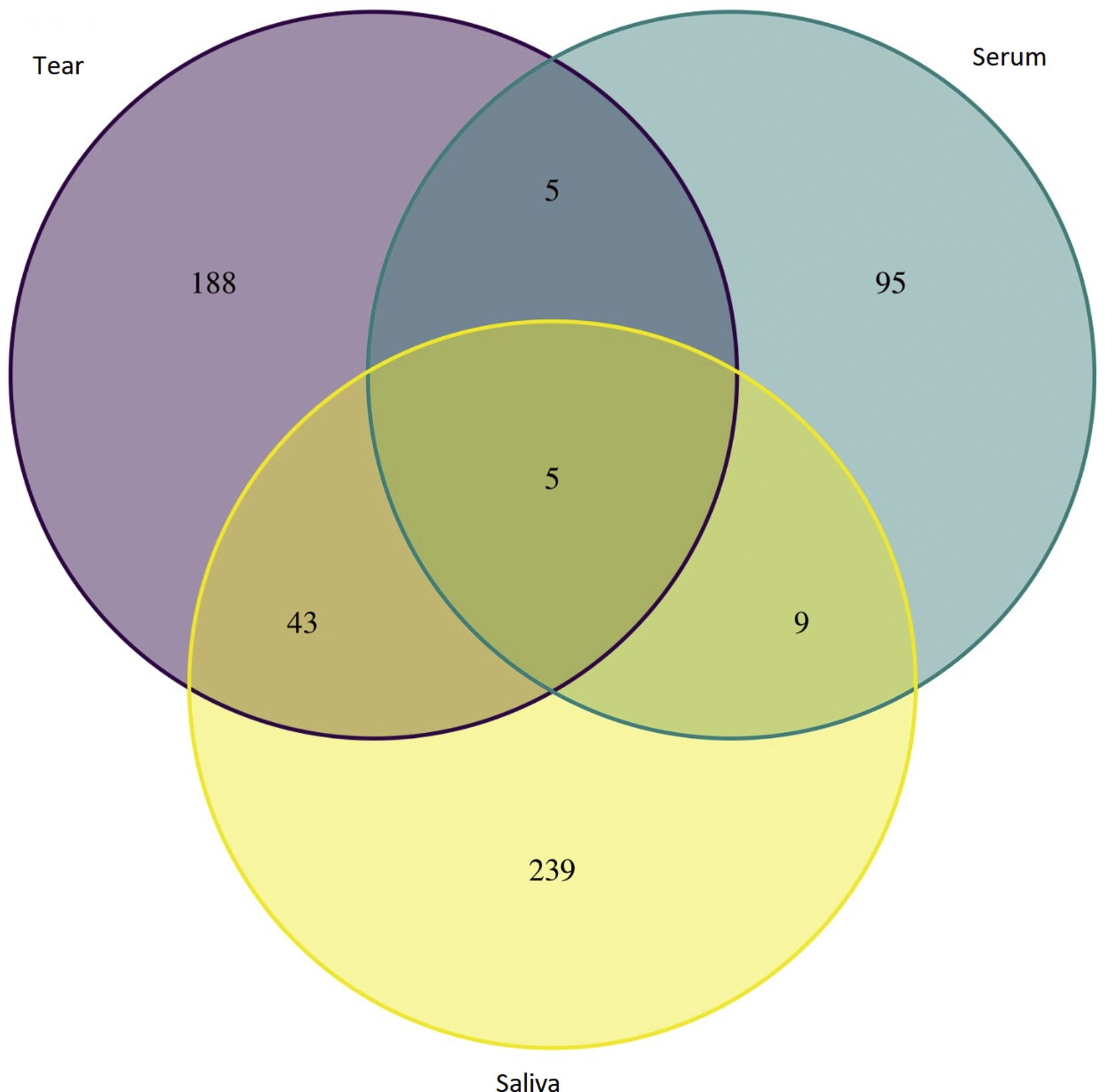
ResultsTen articles met the selection criteria (excluding studies that reported animal or bacterial data),6–17 in total 2968 proteins were analyzed (Table 1). In control group were found 2210 saliva proteins and SS group were found 758 proteins, which were classified into 3 groups saliva (396 proteins), tear (241 proteins) and serum (121 proteins).
An initial pairing using RStudio was done, in which 5 proteins were found in all Sjögren patients and were not in control groups (Table 2). Next, all proteins from SS patients were mapped using REACTOME 7.O, Table 3 shows enriched pathways.
| Intersection | Enriched pathways | p-Value |
|---|---|---|
| Saliva/tear/serum | Extracellular serine/threonine protein kinase FAM20C | 8.75E−5 |
| Saliva/tear/serum | Regulation of insulin-like growth factor (IGF) transport and uptake by insulin-like growth factor binding proteins (IGFBPs) | 3.97E−4 |
| Saliva/tear/serum | Neutrophil degranulation | 7.45E−3 |
Enriched pathways. The proteins were mapped using REACTOME 7.O.
Clinical diagnosis and follow-up of dry eye patients is a great challenge to health professionals because there seems to be no clear correlation between symptoms and results of clinical tests,5 which highlights the necessity of new diagnosis methods.7–17 This study aims to find proteins that can be used as biomarkers in SS, using an adaptation to Ghojavand et al. protocol, that tends to generally integrate reported data to explore events in a holistic way.12
Among the proteins that were found two caught our attention: antithrombin III and E-cadherin, since the increase of its expression is related to SS. Antithrombin III is a glycoprotein dependent on liver synthesized vitamin k, which acts as natural inhibitor of coagulation.18 Antithrombin III inhibits multiple points of the Coagulation cascade, and because of this, its deficiency is associated with thrombosis.18 Of all acquired thrombophilias, antiphospholipid syndrome is the most frequent with 28% of the cases, and it has been associated with other pathologies like Sjögren, rheumatoid arthritis, vasculitis and Crohn's disease.18,19
E-cadherin is an adhesion molecule normally expressed on epithelial cells, required for the formation of adherence junctions between mature epithelial cells, regulating differentiation, proliferation and apoptosis high levels of E-cadherin in serum have been associated with Sjögren and skin inflammation.20 To our surprise high levels of E-cadherin were also reported in saliva and tears of SS patients which makes us suggest the use of this protein as a potential biomarker.
Of the enriched pathways, neutrophil degranulation pathway has been related in many occasions with SS,21 which make us think that it could be related with E-cadherin expression that affects polarization in immune cells, thus driving cell signaling out of control.22
The other two pathways are indirectly associated with autoimmune diseases. Extracellular serine/threonine protein kinase FAM20C that phosphorylates secretory pathway and plays a key role in biomineralization of bones and teeth. In mouse was demonstrate that the Emk protein kinase is essential for maintaining immune system homeostasis and that loss of Emk may contribute to autoimmune disease in mammals.23
The IGF/IGF pathway plays important and diverse roles in tissue development and function. It regulates cell cycle progression, apoptosis, and the translation of proteins. This signaling may also participate in the pathogenesis of autoimmune diseases, although its relationship with these processes seems complex and relatively unexplored, but suggests that this pathway might constitute an attractive therapeutic target.24
Finally, this study suggests antithrombin III and E-cadherin. as biomarkers in SS, because these proteins are differentially expressed in plasma, blood and tear, and suggests neutrophil degranulation pathway as a target for SS.
Conflict of interestThe authors declare that they have no conflict of interest.