The present study aimed to contribute to the catalog of genetic mutations involved in the carcinogenic processes of uterine sarcomas (USs) and carcinosarcomas (UCSs), which may assist in the accurate diagnosis of, and selection of treatment regimens for, these conditions.
METHODS:We performed gene-targeted next-generation sequencing (NGS) of 409 cancer-related genes in 15 US (7 uterine leiomyosarcoma [ULMS], 7 endometrial stromal sarcoma [ESS], 1 adenosarcoma [ADS]), 5 UCS, and 3 uterine leiomyoma (ULM) samples. Quality, frequency, and functional filters were applied to select putative somatic variants.
RESULTS:Among the 23 samples evaluated in this study, 42 loss-of-function (LOF) mutations and 111 missense mutations were detected, with a total of 153 mutations. Among them, 66 mutations were observed in the Catalogue of Somatic Mutations in Cancer (COSMIC) database. TP53 (48%), ATM (22%), and PIK3CA (17%) were the most frequently mutated genes. With respect to specific tumor subtypes, ESS showed mutations in the PDE4DIP, IGTA10, and DST genes, UCS exhibited mutations in ERBB4, and ULMS showed exclusive alterations in NOTCH2 and HER2. Mutations in the KMT2A gene were observed exclusively in ULM and ULMS. In silico pathway analyses demonstrated that many genes mutated in ULMS and ESS have functions associated with the cellular response to hypoxia and cellular response to peptide hormone stimulus. In UCS and ADS, genes with most alterations have functions associated with phosphatidylinositol kinase activity and glycerophospholipid metabolic process.
CONCLUSION:This preliminary study observed pathogenic mutations in US and UCS samples. Further studies with a larger cohort and functional analyses will foster the development of a precision medicine-based approach for the treatment of US and UCS.
Sarcomas are rare heterogeneous tumors that affect the female genital tract and originate from tissues such as muscle, fat, bones, and fibrous tissue. Uterine sarcomas (USs) are the most commonly occurring gynecological sarcomas, representing 90% of the total cases (1). Based on their histological composition, uterine tumors with mesenchymal elements can be divided into 1) pure sarcomas (uterine leiomyosarcomas - ULMSs, endometrial stromal sarcomas - ESSs); 2) mixed epithelial and mesenchymal tumors (adenosarcomas - ADSs), and 3) carcinosarcomas - UCSs, a biphasic tumor composed of high-grade carcinomatous and sarcomatous components derived from transdifferentiation of carcinoma (2). Many studies have characterized UCS tumors as mixed USs; however, since 2014, they have been reclassified as endometrial carcinomas (ECs) that demonstrate metaplastic features (3,4). Despite their low prevalence, USs are associated with high rates of local recurrence, distant metastases, and poor prognosis, with two-year survival rates below 50% (1).
Several genetic alterations have been associated with USs and UCSs, with few alterations being associated with specific histological subtypes. For instance, ESSs can be divided into two types: low-grade ESS (LG-ESS) and high-grade ESS (HG-ESS), both characterized by recurrent chromosomal translocations. In LG-ESS, the most common translocation, t [7; 17] (p15; q21), is observed in almost 50% of the cases and results in the JAZF1-SUZ12 gene fusion (5). Ma et al. (6), revealed that the JAZF1-SUZ12 fusion protein destabilizes polycomb repressive complex 2 (PRC2), abolishes histone methyltransferase (HMT) activity, and subsequently activates genes normally repressed by PRC2. JAZF1-PHF1, EPC1-PHF1, PHF1-MEAF6, MBTD1-CXorf67, and JAZF1-BCORL1 are other less frequent fusion proteins observed in the patients with these tumors. HG-ESS exhibits a YWHAE-NUTM2 gene rearrangement (previously termed YWHAE-FAM22). Recently, molecular alterations in ZC3H7B-BCOR, BCOR-ITD, EPC1-BCOR, JAZF1-BCORL1, and BRD8-PHF1 have been identified. This histological subtype demonstrates more aggressive clinical behavior and worse prognosis (5,2). Many previous studies have investigated the ESS genome with a focus on genetic fusions (7–10). However, Choi et al. (11) demonstrated that fusions are not the only genetic alterations that occur during the development of ESS. Using whole-exome sequencing methods, the aforementioned study described mutations in PTEN, RB1, TP53, and CDH1. Despite the use of a very small number of ESS samples in this study (3 LG-ESS), it is a valuable contribution to the understanding of the pathogenesis of such tumors.
ULMSs are not characterized by specific chromosome translocations; however, they are associated with a complex karyotype with chromosomal gains and losses, such as deletion in chromosome 1. Most ULMSs express PDGFR-α, WT1, CYP19, and GNRH-R (12,13). Owing to gene alterations, the loss of function in the tumor suppressor genes, BRCA1 and MED12 as well as the loss of expression of the proteasome β1i subunit LMP2 have been associated with ULMS development (14). Additionally, The Cancer Genome Atlas (TCGA) Research Network (15) examined the molecular characterization of adult soft tissue sarcomas (STSs) and observed that ULMSs shared more similarities with extrauterine LMSs than that with other sarcomas. Although both tumors exhibit the same pattern of cell differentiation, their tumor environments are extremely diverse. This study included 53 cases of soft-tissue LMS (extrauterine) and 27 ULMS cases that were evaluated by whole-exome sequencing, demonstrating frequent alterations in TP53, RB1, ATRX, and MED12 (16).
Somatic mutations have also been described occurring at low frequency in the majority of the tyrosine kinase growth factor gene family and their targets, namely, v-raf murine sarcoma viral oncogene homolog B1 (BRAF), CDKN2A, epidermal growth factor receptor (EGFR), HER2, v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog (KIT), v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS), platelet-derived growth factor receptor (PDGFR), and phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit α (PI3KCA) during the development of UCS. In addition, mutations in TP53, PTEN, protein phosphatase 2 scaffold subunit alpha (PPP2R1A), F-box, and WD repeat domain containing 7 (FBXW7) have already been identified, which may contribute to the development of therapeutic alternatives including the use of the inhibitors of PARP, EZH2, cell-cycle, and PI3K pathway (14,17). Little information is available on how mutations contribute to ADS etiology; however, one study observed that DICER1 mutations are associated with the tumorigenic process in a small subset of such tumors (18).
Since these are rare tumors, only a few studies focusing on the definition of the mutational repertoire of the different histological types of rare sarcomas have been conducted thus far. Therefore, studies focusing on the mutational characterization of these tumors are of paramount importance and will contribute to the discovery of new biomarkers for precision medicine-based approaches in the treatment of such neoplasms. Herein, we investigated the mutational profile of the samples obtained from patients with ULMSs, UCSs, ESSs, and ADSs, using a commercial panel containing 409 cancer-associated genes involved in apoptosis, signaling, transcription regulation, inflammation response, and growth factors-associated pathway.
MATERIALS AND METHODSSample selectionIn order to analyze differences in genetic mutations between different histological types of US, we initially selected 43 formalin-fixed and paraffin-embedded (FFPE) human samples including 14 ULMS, 12 ESS, 2 ADS, 12 UCS, and 3 ULM-non-cancerous tumor (as reference samples). All samples were obtained via surgical procedures performed between 2000 and 2012 at the Institute of Cancer of Sao Paulo (ICESP) and Clinics Hospital of the Faculty of Medicine, University of Sao Paulo (HCFMUSP). Tissues were stored at the molecular and structural gynecology laboratory (LIM-58) of the University of Sao Paulo Medical School (FMUSP).
This study was performed in accordance with the Declaration of Helsinki and was approved by the Research Ethics Committee of the FMUSP with protocol number 477/15. Patients’ medical records were revised and the following data were recorded: age at diagnosis, postmenopausal bleeding, adjuvant treatment, presence of metastasis or recurrence, and status.
DNA IsolationGenomic DNA was extracted using the QIAamp DNA FFPE Tissue Kit obtained from QIAGEN® according to the manufacturer's instructions. DNA concentration, purity, and integrity were assessed by spectrophotometry (Nanodrop 2000, Thermo Fisher Scientific) and fluorometry (Qubit - Thermo Fisher Scientific), respectively.
Preparation of sequencing libraries and Next-Generation Sequencing (NGS)Sequencing libraries were prepared using the Ion Torrent Ampliseq Comprehensive Cancer Panel - Catalog number: 4477685 (Thermo Fisher Scientific), which contains ∼16,000 primer pairs multiplexed into 4 pools. This commercial panel was designed to assess the mutational profile of 409 cancer driver genes and drug targets along with signaling cascades, apoptosis genes, DNA repair genes, transcription regulators, inflammatory response genes, and growth factor genes (Table S1). Prior to amplification, DNA was treated with the uracil-DNA glycosylase enzyme (Thermo Fisher Scientific) by adding 1 unit of enzyme per 50 ng of DNA and incubating for 15 min at 37 °C. This procedure was performed to remove DNA molecules containing uracil and decrease the number of artifactual variants in the sequencing (19). Libraries were then prepared using Ion AmpliSeqTM Library kit 2.0 protocols, with 10 ng of input DNA per pool, totaling 40 ng of DNA from each sample. The FuPa reagent was used to partially digest primer sequences and phosphorylate the amplicons. Next, sequencing adaptors and barcodes were ligated to the amplicon by the enzyme Ligase using the Ion Xpress™ Barcode Adapters kit (Thermo Fisher Scientific), which were then purified using magnetic beads (Agencourt® AMPure® XP Reagents, Beckman Coulter). Subsequently, emulsion PCR was performed using the Ion PI™ Hi-Q™ OT2 200 Kit (Thermo Fisher Scientific), followed by sequencing with Ion PITM Hi-QTM sequencing 200 and Ion PITM Chip.
Ion AmpliSeq Comprehensive Cancer Panel target gene list.
| 1 | ABL1 | CBL | EP300 | GATA2 | LAMP1 | MYD88 | PKHD1 | SMARCA4 | WHSC1 |
| 2 | ABL2 | CCND1 | EP400 | GATA3 | LCK | MYH11 | PLAG1 | SMARCB1 | WRN |
| 3 | ACVR2A | CCND2 | EPHA3 | GDNF | LIFR | MYH9 | PLCG1 | SMO | WT1 |
| 4 | ADAMTS20 | CCNE1 | EPHA7 | GNA11 | LPHN3 | NBN | PLEKHG5 | SMUG1 | XPA |
| 5 | AFF1 | CD79A | EPHB1 | GNAQ | POT1 | NCOA1 | PML | SOCS1 | XPC |
| 6 | AFF3 | CD79B | EPHB4 | GNAS | LPP | NCOA2 | PMS1 | SOX11 | XPO1 |
| 7 | AKAP9 | CDC73 | EPHB6 | GPR124 | LRP1B | NCOA4 | PMS2 | SOX2 | XRCC2 |
| 8 | AKT1 | CDH1 | ERBB2 | GRM8 | LTF | NF1 | POU5F1 | SRC | ZNF384 |
| 9 | AKT2 | CDH11 | ERBB3 | GUCY1A2 | LTK | NF2 | PPARG | SSX1 | ZNF521 |
| 10 | AKT3 | CDH2 | ERBB4 | HCAR1 | MAF | NFE2L2 | PPP2R1A | STK11 | |
| 11 | ALK | CDH20 | ERCC1 | HIF1A | MAFB | NFKB1 | PRDM1 | STK36 | |
| 12 | APC | CDH5 | ERCC2 | HLF | MAGEA1 | NFKB2 | PRKAR1A | SUFU | |
| 13 | AR | CDK12 | ERCC3 | HNF1A | MAGI1 | NIN | PRKDC | SYK | |
| 14 | ARID1A | CDK4 | ERCC4 | HOOK3 | MALT1 | NKX2-1 | PSIP1 | SYNE1 | |
| 15 | ARID2 | CDK6 | ERCC5 | HRAS | MAML2 | NLRP1 | PTCH1 | TAF1 | |
| 16 | ARNT | CDK8 | ERG | HSP90AA1 | MAP2K1 | NOTCH1 | PTEN | TAF1L | |
| 17 | ASXL1 | CDKN2A | ESR1 | HSP90AB1 | MAP2K2 | NOTCH2 | PTGS2 | TAL1 | |
| 18 | ATF1 | CDKN2B | ETS1 | ICK | MAP2K4 | NOTCH4 | PTPN11 | TBX22 | |
| 19 | ATM | CDKN2C | ETV1 | IDH1 | MAP3K7 | NPM1 | PTPRD | TCF12 | |
| 20 | ATR | CEBPA | ETV4 | IDH2 | MAPK1 | NRAS | PTPRT | TCF3 | |
| 21 | ATRX | CHEK1 | EXT1 | IGF1R | MAPK8 | NSD1 | RAD50 | TCF7L1 | |
| 22 | AURKA | CHEK2 | EXT2 | IGF2 | MARK1 | NTRK1 | RAF1 | TCF7L2 | |
| 23 | AURKB | CIC | EZH2 | IGF2R | MARK4 | NTRK3 | RALGDS | TCL1A | |
| 24 | AURKC | CKS1B | FAM123B | IKBKB | MBD1 | NUMA1 | RARA | TET1 | |
| 25 | AXL | CMPK1 | FANCA | IKBKE | MCL1 | NUP214 | RB1 | TET2 | |
| 26 | BAI3 | COL1A1 | FANCC | IKZF1 | MDM2 | NUP98 | RECQL4 | TFE3 | |
| 27 | BAP1 | CRBN | FANCD2 | IL2 | MDM4 | PAK3 | REL | TGFBR2 | |
| 28 | BCL10 | CREB1 | FANCF | IL21R | MEN1 | PALB2 | RET | TGM7 | |
| 29 | BCL11A | CREBBP | FANCG | IL6ST | MET | PARP1 | RHOH | THBS1 | |
| 30 | BCL11B | CRKL | FAS | IL7R | MITF | PAX3 | RNASEL | TIMP3 | |
| 31 | BCL2 | CRTC1 | FBXW7 | ING4 | MLH1 | PAX5 | RNF2 | TLR4 | |
| 32 | BCL2L1 | CSF1R | FGFR1 | IRF4 | MLL | PAX7 | RNF213 | TLX1 | |
| 33 | BCL2L2 | CSMD3 | FGFR2 | IRS2 | MLL2 | PAX8 | ROS1 | TNFAIP3 | |
| 34 | BCL3 | CTNNA1 | FGFR3 | ITGA10 | MLL3 | PBRM1 | RPS6KA2 | TNFRSF14 | |
| 35 | BCL6 | CTNNB1 | FGFR4 | ITGA9 | MLLT10 | PBX1 | RRM1 | TNK2 | |
| 36 | BCL9 | CYLD | FH | ITGB2 | MMP2 | PDE4DIP | RUNX1 | TOP1 | |
| 37 | BCR | CYP2C19 | FLCN | ITGB3 | MN1 | PDGFB | RUNX1T1 | TP53 | |
| 38 | BIRC2 | CYP2D6 | FLI1 | JAK1 | MPL | PDGFRA | SAMD9 | TPR | |
| 39 | BIRC3 | DAXX | FLT1 | JAK2 | MRE11A | PDGFRB | SBDS | TRIM24 | |
| 40 | BIRC5 | DCC | FLT3 | JAK3 | MSH2 | PER1 | SDHA | TRIM33 | |
| 41 | BLM | DDB2 | FLT4 | JUN | MSH6 | PGAP3 | SDHB | TRIP11 | |
| 42 | BLNK | DDIT3 | FN1 | KAT6A | MTOR | PHOX2B | SDHC | TRRAP | |
| 43 | BMPR1A | DDR2 | FOXL2 | KAT6B | MTR | PIK3C2B | SDHD | TSC1 | |
| 44 | BRAF | DEK | FOXO1 | KDM5C | MTRR | PIK3CA | SEPT9 | TSC2 | |
| 45 | BRD3 | DICER1 | FOXO3 | KDM6A | MUC1 | PIK3CB | SETD2 | TSHR | |
| 46 | BRIP1 | DNMT3A | FOXP1 | KDR | MUTYH | PIK3CD | SF3B1 | UBR5 | |
| 47 | BTK | DPYD | FOXP4 | KEAP1 | MYB | PIK3CG | SGK1 | UGT1A1 | |
| 48 | BUB1B | DST | FZR1 | KIT | MYC | PIK3R1 | SH2D1A | USP9X | |
| 49 | CARD11 | EGFR | G6PD | KLF6 | MYCL1 | PIK3R2 | SMAD2 | VHL | |
| 50 | CASC5 | EML4 | GATA1 | KRAS | MYCN | PIM1 | SMAD4 | WAS |
The results were analyzed using the Torrent Suite v5.0.5 software (Thermo Fisher Scientific). Sequence variants (SNVs and indels) were identified using the Torrent Variant Caller (Ion Torrent - Thermo Fisher Scientific) and compared to the GRCh37 / hg19 genome version. VCF files were analyzed using VarSeq v1.8 software (GoldenHelix) for variant annotation and prioritization. The variants were filtered based on the quality and frequency criteria: coverage (>100), genotype quality score cutoff (GQS>50), variant base in at least 5% of reads, variant base present in at least 2 reads in each direction, homopolymer-length error<5, absence of genetic variants in population databases (ExAC; NHLBI-ESP; 1000 Genomes Project) or minor allele frequency (MAF)≤0.01%.
Subsequently, variants were selected based on their effect on protein expression, with the following being considered: 1) variants described in the COSMIC database; 2) loss-of-function variants - Frameshift variants-nucleotide insertions/deletions, gain/loss of stop codons, splice site alterations); or 3) missense variants (in-frame insertions/deletions, amino acid exchange) predicted as possibly pathogenic in at least three of six prediction programs used (SIFT, PolyPhen, MutationTaster, MutationAssessor, FATHMM, FATHMM-MKL) and occurring in oncogenes or tumor suppressor genes in OncoMD database. Variants not previously described in the COSMIC database were visually inspected using the integrative genomics viewer (IGV) program to exclude sequencing artifacts.
Construction of genetic interaction networks was performed using Cytoscape platform version 3.7.0, which uses data from protein and genetic interactions, pathways, co-expression, co-localization, and protein domain similarity.
RESULTSInitially, 40 US and UCS (14 ULMS, 12 ESS, 2 ADS, and 12 UCS) and 3 ULM samples were selected from the pathology department files; however, only 23 (7 ULMS, 7 ESS, 1 ADS, 5 UCS, and 3 ULM) remained until the end of NGS analyses. Some losses occurred while performing multiplex PCR reactions (AmpliSeq™), during which we observed a high degree of fragmented DNA and many genetic artifacts in several samples. These issues are expected since tissue processing for paraffin inclusion and long storage time causes damage to the DNA structure (integrity). The clinical and pathological features of 40 patients with US and UCS who were enrolled in this study are summarized in Table 1.
Clinical and pathological features of US and UCS patients (n=40).
| Variables | Categories | US/UCS n (%) |
|---|---|---|
| Age | >50 years | 33 (82) |
| ≤50 years | 7 (18) | |
| N.A. | 0 (0) | |
| Postmenopausal Bleeding | Yes | 22 (55) |
| No | 13 (33) | |
| N.A. | 5 (12) | |
| Adjuvant Treatment | No | 8 (20) |
| RT | 19 (47) | |
| CT | 8 (20) | |
| RT+CT | 5 (13) | |
| N.A. | 0 (0) | |
| Metastasis or Recurrence | Yes | 22 (55) |
| No | 14 (35) | |
| N.A. | 4 (10) | |
| Status | Alive | 11 (27) |
| Death | 23 (58) | |
| Loss of follow-up | 6 (15) | |
| N.A. | 0 (0) |
radiotherapy (RT); chemotherapy (CT); not available (NA); uterine sarcomas (US).
*ULM samples were not included owing to their benign characterization.
Among the 23 samples deemed suitable for the evaluation of sequencing data, homogeneity average was 73.2%, median base coverage was 1257X, and horizontal coverage was 84.3% corresponding to 100X. Based on the NGS data, we selected point mutations with possible impacts on the function of the protein encoded by the altered gene (missense, nonsense, splice-site mutations, loss of stop codons) and small insertions and deletions (indels). Total variants detected in each sample and filtered variants for the selection of somatic alterations of interest are presented in Table 2.
Total variants obtained after filtering performed to increase the specificity of NGS results (higher stringency).
| General (pre-filters) | Selected Variants | ||||||
|---|---|---|---|---|---|---|---|
| Samples | Total | SNV | Insertions | Deletions | LOFs | Missense | Cosmic |
| ESS 2 | 2347 | 2257 | 40 | 50 | 1 | 6 | 5 |
| ESS 3 | 1551 | 1473 | 31 | 47 | 2 | 6 | 4 |
| ESS 4 | 1249 | 1162 | 36 | 51 | 1 | 1 | 1 |
| ESS 5 | 1416 | 1324 | 36 | 56 | 0 | 4 | 3 |
| ESS 7 | 1494 | 1397 | 40 | 57 | 2 | 2 | 1 |
| ESS 9 | 1421 | 1343 | 35 | 43 | 1 | 6 | 5 |
| ESS 10 | 1440 | 1329 | 35 | 76 | 4 | 6 | 6 |
| UCS 2 | 1332 | 1223 | 47 | 62 | 3 | 4 | 4 |
| UCS 5 | 1362 | 1271 | 42 | 49 | 7 | 13 | 7 |
| UCS 9 | 1972 | 1884 | 42 | 46 | 1 | 6 | 4 |
| UCS 13 | 1234 | 1150 | 36 | 48 | 1 | 6 | 4 |
| UCS 19 | 1604 | 1516 | 33 | 55 | 1 | 3 | 3 |
| ULMS 38 | 746 | 678 | 43 | 25 | 0 | 7 | 4 |
| ULMS 39 | 1768 | 1688 | 34 | 46 | 1 | 2 | 2 |
| ULMS 40 | 1296 | 1193 | 42 | 61 | 2 | 3 | 1 |
| ULMS 45 | 2004 | 1921 | 36 | 47 | 2 | 3 | 1 |
| ULMS 52 | 2806 | 2746 | 23 | 37 | 0 | 11 | 6 |
| ULMS 50 | 2842 | 2651 | 77 | 114 | 0 | 1 | 0 |
| ULMS 59 | 2132 | 1968 | 76 | 88 | 4 | 6 | 2 |
| ADS 2 | 3521 | 3406 | 41 | 74 | 6 | 10 | 0 |
| ULM 119 | 1298 | 1201 | 37 | 60 | 1 | 1 | 0 |
| ULM 143 | 981 | 919 | 33 | 29 | 1 | 2 | 2 |
| ULM 152 | 1297 | 1237 | 25 | 35 | 1 | 2 | 1 |
*Endometrial stromal sarcoma (ESS); Uterine carcinosarcoma (UCS); Uterine leiomyosarcoma (ULMS); Adenocarcinoma (ADS); Uterine leiomyoma (ULM).Single nucleotide variant (SNV); Loss of function (LOFs); Catalogue of Somatic Mutations in Cancer (COSMIC).
An average of 1700 alterations were identified per sample (ranging from 746 to 3521), with an average of 1606 single nucleotide variants (SNVs) (ranging from 678 to 3406), 40 insertions (ranging from 23 to 77), and 55 deletions (ranging from 25 to 114). To select relevant somatic variants, a first filter was applied focusing on the quality and frequencies of these alterations. A second filter, focusing on variant functions and effects, was used to select the alterations that would be most relevant in alterations of gene functions. Collectively, in 23 samples that were evaluated, 42 LOF mutations and 111 missense mutations were detected, with a total of 153 filtered mutations, among which 66 were found in the COSMIC database (Table 2).
Among the 409 genes included in the panel, mutations were detected in 94 distinct genes, with 30 genes demonstrating mutations in more than one sample and 64 genes showing mutations in a single sample. Table 3 presents the list of genes that were mutated in more than one sample of the cohort, along with the number of mutated samples and the histological types. TP53 (11/23 - 48%), ATM (5/23 - 22%), and PIK3CA (4/23 - 17%) were the most frequently mutated genes.
Gene mutations observed in more than one sample and histological subtypes.
| Gene | Mutated samples n (%) | Histological Types (ULMS/ESS/UCS/ADS/ULM) |
|---|---|---|
| TP53 | 11 (48%) | 4 ULMS, 3 ESS, 4 UCS |
| ATM | 5 (22%) | 2 ULMS, 2 ESS, 1 UCS |
| PIK3CA | 4 (17%) | 1 ESS, 3 UCS |
| KMT2D | 3 (13%) | 1 ULMS, 1 ESS, 1 UCS |
| MTOR | 3 (13%) | 1 ESS, 1 UCS, 1 ULM |
| JAK3 | 3 (13%) | 1 ULMS, 1 ESS, 1 ULM |
| APC | 3 (13%) | 1 ULMS, 2 ESS |
| DICER1 | 3 (13%) | 1 ESS, 2 UCS |
| TRRAP | 3 (13%) | 2 UCS, 1 ADS |
| TSC2 | 3 (13%) | 2 ULMS, 1 ADS |
| PDE4DIP | 3 (13%) | 3 ESS |
| AR | 2 (9%) | 1 ESS, 1 UCS |
| ATRX | 2 (9%) | 1 ULMS, 1 ESS |
| CREBBP | 2 (9%) | 1 ULMS, 1 ESS |
| DNMT3A | 2 (9%) | 1 UCS, 1 ADS |
| EPHA7 | 2 (9%) | 1 UCS, 1 ADS |
| KAT6B | 2 (9%) | 1 UCS, 1 ADS |
| KMT2A | 2 (9%) | 1 ULMS, 1 ULM |
| MET | 2 (9%) | 1 UCS, 1 ULM |
| MYB | 2 (9%) | 1 ULMS, 1 ESS |
| NOTCH1 | 2 (9%) | 1 ULMS, 1 UCS |
| PRKDC | 2 (9%) | 1 UCS, 1 ADS |
| SYNE1 | 2 (9%) | 1 ULMS, 1 ESS |
| NF1 | 2 (9%) | 1 ESS, 1 UCS |
| NOTCH2 | 2 (9%) | 2 ULMS |
| HER2 | 2 (9%) | 2 ULMS |
| ERBB4 | 2 (9%) | 2 UCS |
| DAXX | 2 (9%) | 1 ESS, 1 ADS |
| ITGA10 | 2 (9%) | 2 ESS |
| DST | 2 (9%) | 2 ESS |
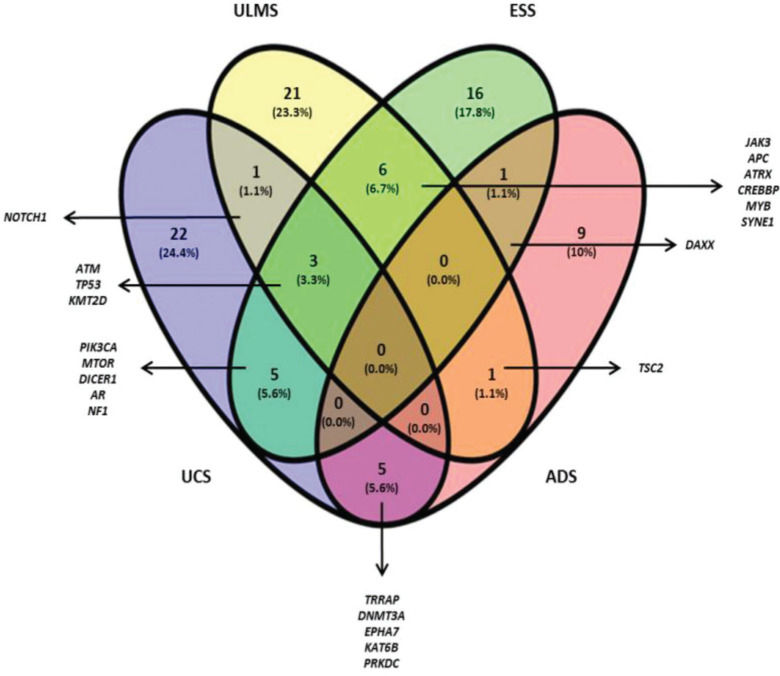
The Venn diagram (Figure 1) shows the shared and individual (specific) mutations of each malignant histological subtype evaluated (ULMS, ESS, UCS, and ADS). Three shared genes were observed (ATM, TP53, and KMT2D) among the ULMS, ESS, and UCS samples. Nineteen genes were shared between 2 types of tumors, and 68 genes were mutated in a single type. Among them, 6 genes were mutated in more than one sample of the same histological subtype, namely, PDE4DIP (3 ESS samples), ITGA10, and DST (2 ESS samples), NOTCH2, and HER2 (2 ULMS samples), and ERBB4 (2 UCS samples). Quantitatively, this analysis shows similarities in the mutational profiles of ULMS and ESS, with 6 mutated genes in common (6.7%) between both subtypes. In the genes JAK3, APC, ATRX, CREBBP, MYB, and SYNE1, most of the mutations were characterized as missense mutations; however, in the SYNE1 gene, the two mutations observed in ULMS and ESS samples were determined as LOF mutations (c.352C>T and c.8565G>A, respectively). In addition, mutations in the TRRAP, DNMT3A, EPHA7, KAT6B, and PRKDC genes indicate that UCS and ADS may exhibit molecular similarities.
Table 4 summarizes the genes with the most frequent alterations (mutations in 2 or more samples, or with 2 mutations in the same sample), the types of mutations, and their position. Alterations in the respective proteins are also indicated, along with the combined effect of these alterations (Missense or LOF) and DNA (c.), and protein (p.) nomenclatures. Their nomenclature can be used for database searches. The descriptions of the 153 potentially somatic variants are listed in Table S2. UCS5, ULMS52, ESS58107, and ADS2 samples demonstrated the highest number of mutations (UCS5 with 20 mutations in 19 genes; ULMS52 with 11 mutations in 10 genes; ESS58107 with 10 mutations in 10 genes, and ADS2 with 16 mutations in 16 genes). Samples with the lowest number of mutations were ULMS50b with 1 mutation in ALK, ESS4 with 2 mutations (ATM and CREBBP), and ULM119 (benign tissue) with 2 mutations (MET and PDGFB).
Most common mutations observed in the study, their chromosomal positions, effects, and nomenclature.
| Sample | Chr:Pos | Gene | HGVS c. | HGVS p. | Effect |
|---|---|---|---|---|---|
| UCS2 | 3:178921549 | PIK3CA | c.1031T>C | p.Val344Ala | Missense |
| 6:94120318 | EPHA7 | c.733G>A | p.Ala245Thr | Missense | |
| 7:116339356 | MET | c.218T>A | p.Leu73Ter | LOF: stop - gained | |
| 8:48776121 | PRKDC | c.5586delT | p.Phe1862Leufs | LOF: frameshift | |
| 17:7577547 | TP53 | c.734G>T | p.Gly245Val | Missense | |
| UCS5 | 1:11227575 | MTOR | c.4254-1G>A | r.spl? | LOF: splice - acceptor |
| 3:178952085 | PIK3CA | c.3140A>G | p.His1047Arg | Missense | |
| 10:76735809 | KAT6B | c.1714C>T | p.Arg572Cys | Missense | |
| 11:108114777 | ATM | c.594C>A | p.Cys198Ter | LOF: stop - gained | |
| 14:95572101 | DICER1 | c.3007C>T | p.Arg1003Ter | LOF: stop - gained | |
| 17:29588751 | NF1 | c.4600C>T | p.Arg1534Ter | LOF: stop - gained | |
| 17:29665110 | NF1 | c.6772C>T | p.Arg2258Ter | LOF: stop - gained | |
| 2:25469168 | DNMT3A | c.1290T>G | p.Asn430Lys | Missense | |
| 2:212587219 | ERBB4 | c.782A>C | p.Gln261Pro | Missense | |
| 7:98513427 | TRRAP | c.2281C>T | p.Arg761Trp | Missense | |
| X:66766207 | AR | c.1219C>T | p.Arg407Cys | Missense | |
| UCS9 | 9:139391355 | NOTCH1 | c.6836C>T | p.Ala2279Val | Missense |
| 12:49444719 | KMT2D | c.2747C>T | p.Pro916Leu | Missense | |
| 17:7578442 | TP53 | c.488A>G | p.Tyr163Cys | Missense | |
| UCS13 | 3:178916854 | PIK3CA | c.241G>A | p.Glu81Lys | Missense |
| 14:95574253 | DICER1 | c.2614G>A | p.Ala872Thr | Missense | |
| 17:7577534 | TP53 | c.747G>T | p.Arg249Ser | Missense | |
| 7:98609947 | TRRAP | c.11549G>A | p.Arg3850His | Missense | |
| UCS19 | 2:212295800 | ERBB4 | c.2513G>A | p.Arg838Gln | Missense |
| 17:7577580 | TP53 | c.701A>G | p.Tyr234Cys | Missense | |
| ULMS38 | 1:120458122 | NOTCH2 | c.7223T>A | p.Leu2408His | Missense |
| 17:37864584 | HER2 | c.236A>C | p.Glu79Ala | Missense | |
| 19:17937659 | JAK3 | c.3268G>A | p.Ala1090Thr | Missense | |
| ULMS39 | 17:7577545 | TP53 | c.736A>G | p.Met246Val | Missense |
| ULMS40 | 11:108139268 | ATM | c.2770C>T | p.Arg924Trp | Missense |
| 17:7577120 | TP53 | c.818G>A | p.Arg273His | Missense | |
| 17:37881117 | HER2 | c.2446C>T | p.Arg816Cys | Missense | |
| X:76891445 | ATRX | c.4660A>T | p.Arg1554Ter | LOF: stop - gained | |
| ULMS45 | 11:108160506 | ATM | c.4414T>G | p.Leu1472Val | Missense |
| 17:7578290 | TP53 | c.560-1G>C | r.spl? | LOF: splice - acceptor | |
| 16:2135281 | TSC2 | c.4620C>A | p.Tyr1540Ter | LOF: stop - gained | |
| ULMS52 | 1:120459251 | NOTCH2 | c.6094C>A | p.His2032Asn | Missense |
| 9:139400980 | NOTCH1 | c.4013C>T | p.Ala1338Val | Missense | |
| 11:118377142 | KMT2A | c.10535C>T | p.Pro3512Leu | Missense | |
| 12:49416396 | KMT2D | c.16315C>T | p.Arg5439Trp | Missense | |
| 16:2130319 | TSC2 | c.3551C>T | p.Ala1184Val | Missense | |
| 16:3779521 | CREBBP | c.5527T>C | p.Cys1843Arg | Missense | |
| 16:3790470 | CREBBP | c.4063G>A | p.Gly1355Arg | Missense | |
| 17:7574017 | TP53 | c.1010G>A | p.Arg337His | Missense | |
| ULMS59 | 5:112173857 | APC | c.2566C>T | p.Arg856Cys | Missense |
| 6:135511289 | MYB | c.331G>A | p.Gly111Ser | Missense | |
| ULMS59 | 6:135539101 | MYB | c.2269C>T | p.Arg757Trp | Missense |
| 6:152832196 | SYNE1 | c.352C>T | p.Arg118Ter | LOF: stop - gained | |
| ULMS59 | 20:57429026 | GNAS | c.706G>A | p.Asp236Asn | Missense |
| 20:57480457 | GNAS | c.2381A>C | p.Lys794Thr | Missense | |
| ESS2 (LG-ESS) | 6:152706896 | SYNE1 | c.8565G>A | p.Trp2855Ter | LOF: stop - gained |
| 11:108175463 | ATM | c.5558A>T | p.Asp1853Val | Missense | |
| 17:7577121 | TP53 | c.817C>T | p.Arg273Cys | Missense | |
| ESS2 | 17:7577139 | TP53 | c.799C>T | p.Arg267Trp | Missense |
| ESS3 | 1:145015874 | PDE4DIP | c.214C>T | p.Arg72Ter | LOF: stop - gained |
| 5:112154777 | APC | c.1048T>C | p.Ser350Pro | Missense | |
| 5:112162855 | APC | c.1459G>A | p.Gly487Arg | Missense | |
| 6:56328464 | DST | c.16429C>T | p.Arg5477Trp | Missense | |
| 12:49418436 | KMT2D | c.15977T>C | p.Leu5326Pro | Missense | |
| 17:7578176 | TP53 | c.672+1G>A | r.spl? | LOF: splice - donor | |
| 17:29556250 | NF1 | c.2617C>T | p.Arg873Cys | Missense | |
| 17:29677234 | NF1 | c.7355G>T | p.Arg2452Leu | Missense | |
| ESS4 | 11:108141990 | ATM | c.2934delT | p.Leu979Cysfs | LOF: frameshift |
| 16:3820773 | CREBBP | c.2678C>T | p.Ser893Leu | Missense | |
| ESS5 | 1:11217330 | MTOR | c.4348T>G | p.Tyr1450Asp | Missense |
| 19:17937659 | JAK3 | c.3268G>A | p.Ala1090Thr | Missense | |
| ESS7 | 6:33287248 | DAXX | c.1885G>A | p.Val629Ile | Missense |
| 14:95590677 | DICER1 | c.1232C>A | p.Ser411Ter | LOF: stop - gained | |
| ESS7 | X:76939115 | ATRX | c.1633C>G | p.Gln545Glu | Missense |
| ESS9 | 1:144906139 | PDE4DIP | c.2494delC | p.Gln832Argfs | LOF - frameshift |
| 1:145536012 | ITGA10 | c.2104G>A | p.Ala702Thr | Missense | |
| 3:178936091 | PIK3CA | c.1633G>A | p.Glu545Lys | Missense | |
| 5:112175711 | APC | c.4420G>A | p.Ala1474Thr | Missense | |
| ESS58107 | 1:145015874 | PDE4DIP | c.214C>T | p.Arg72Ter | LOF: stop - gained |
| 1:145536012 | ITGA10 | c.2104G>A | p.Ala702Thr | Missense | |
| 6:56328464 | DST | c.16429C>T | p.Arg5477Trp | Missense | |
| 6:135516944 | MYB | c.1007C>T | p.Thr336Ile | Missense | |
| 17:7578176 | TP53 | c.672+1G>A | r.spl? | LOF: splice - donor | |
| X:66863156 | AR | c.1675A>T | p.Thr559Ser | Missense | |
| ADS2 | 2:25467477 | DNMT3A | c.1599C>A | p.Tyr533Ter | LOF: stop - gained |
| 6:33288629 | DAXX | c.959A>G | p.Gln320Arg | Missense | |
| 6:93979315 | EPHA7 | c.1513C>A | p.Leu505Met | Missense | |
| 7:98501128 | TRRAP | c.1024G>T | p.Glu342Ter | LOF: stop - gained | |
| 8:48711786 | PRKDC | c.10279G>T | p.Glu3427Ter | LOF: stop - gained | |
| 10:76781925 | KAT6B | c.3308_3310delAAG | p.Glu1104del | LOF: inframe/del | |
| 16:2138078 | TSC2 | c.5098G>T | p.Ala1700Ser | Missense | |
| ULM119 | 7:116403114 | MET | c.2429A>C | p.His810Pro | Missense |
| ULM143 | 1:11307996 | MTOR | c.995_996dupGG | p.Leu333Glyfs | LOF: frameshift |
| 19:17945696 | JAK3 | c.2164G>A | p.Val722Ile | Missense | |
| ULM152 | 11:118344893 | KMT2A | c.3019G>T | p.Gly1007Cys | Missense |
Description of 153 potential somatic variants selected in 23 samples of uterine tumors.
| Sample | Chr:Pos | Gene | HGVS c. | HGVS p. | Effect |
|---|---|---|---|---|---|
| UCS2 | 3:178921549 | PIK3CA | c.1031T>C | p.Val344Ala | Missense |
| 6:94120318 | EPHA7 | c.733G>A | p.Ala245Thr | Missense | |
| 7:116339356 | MET | c.218T>A | p.Leu73Ter | LOF: stop - gained | |
| 8:48776121 | PRKDC | c.5586delT | p.Phe1862Leufs | LOF: frameshift | |
| 15:99500303 | IGF1R | c.3736C>T | p.Arg1246Cys | Missense | |
| 17:7577547 | TP53 | c.734G>T | p.Gly245Val | Missense | |
| 22:33253291 | TIMP3 | c.260delC | p.Glu88Argfs | LOF: frameshift | |
| UCS5 | 1:11227575 | MTOR | c.4254-1G>A | r.spl? | LOF: splice - acceptor |
| 1:27105553 | ARID1A | c.5164C>T | p.Arg1722Ter | LOF: stop - gained | |
| 1:65310574 | JAK1 | c.2116-2A>G | r.spl? | LOF: splice - acceptor | |
| 3:178952085 | PIK3CA | c.3140A>G | p.His1047Arg | Missense | |
| 10:76735809 | KAT6B | c.1714C>T | p.Arg572Cys | Missense | |
| 10:97969609 | BLNK | c.731C>T | p.Pro244Leu | Missense | |
| 11:108114777 | ATM | c.594C>A | p.Cys198Ter | LOF: stop - gained | |
| 14:95572101 | DICER1 | c.3007C>T | p.Arg1003Ter | LOF: stop - gained | |
| 17:29588751 | NF1 | c.4600C>T | p.Arg1534Ter | LOF: stop - gained | |
| 17:29665110 | NF1 | c.6772C>T | p.Arg2258Ter | LOF: stop - gained | |
| 19:45260400 | BCL3 | c.646C>T | p.Arg216Cys | Missense | |
| 1:47685756 | TAL1 | c.632G>A | p.Arg211His | Missense | |
| 2:25469168 | DNMT3A | c.1290T>G | p.Asn430Lys | Missense | |
| 2:212587219 | ERBB4 | c.782A>C | p.Gln261Pro | Missense | |
| 7:98513427 | TRRAP | c.2281C>T | p.Arg761Trp | Missense | |
| 9:37015073 | PAX5 | c.331G>A | p.Ala111Thr | Missense | |
| 19:11098401 | SMARCA4 | c.919C>T | p.Pro307Ser | Missense | |
| 20:36030940 | SRC | c.1219G>A | p.Asp407Asn | Missense | |
| X:44942716 | KDM6A | c.3452A>G | p.Gln1151Arg | Missense | |
| X:66766207 | AR | c.1219C>T | p.Arg407Cys | Missense | |
| UCS9 | 9:139391355 | NOTCH1 | c.6836C>T | p.Ala2279Val | Missense |
| 10:123298226 | FGFR2 | c.628C>T | p.Arg210Ter | LOF: stop - gained | |
| 12:49444719 | KMT2D | c.2747C>T | p.Pro916Leu | Missense | |
| 15:40916649 | KNL1 | c.4265G>A | p.Arg1422Gln | Missense | |
| 17:7578442 | TP53 | c.488A>G | p.Tyr163Cys | Missense | |
| 3:52440867 | BAP1 | c.637C>T | p.Arg213Cys | Missense | |
| 21:39755729 | ERG | c.1057G>A | p.Glu353Lys | Missense | |
| UCS13 | 3:178916854 | PIK3CA | c.241G>A | p.Glu81Lys | Missense |
| 11:71726283 | NUMA1 | c.2266G>T | p.Glu756Ter | LOF: stop - gained | |
| 13:29001422 | FLT1 | c.1310C>T | p.Ser437Leu | Missense | |
| 14:95574253 | DICER1 | c.2614G>A | p.Ala872Thr | Missense | |
| 17:7577534 | TP53 | c.747G>T | p.Arg249Ser | Missense | |
| 5:176636902 | NSD1 | c.1502A>G | p.Lys501Arg | Missense | |
| 7:98609947 | TRRAP | c.11549G>A | p.Arg3850His | Missense | |
| UCS19 | 2:212295800 | ERBB4 | c.2513G>A | p.Arg838Gln | Missense |
| 9:5126715 | JAK2 | c.3323A>G | p.Asn1108Ser | Missense | |
| 17:7577580 | TP53 | c.701A>G | p.Tyr234Cys | Missense | |
| 17:37829120 | PGAP3 | c.900-1G>A | r.spl? | LOF: splice - acceptor | |
| ULMS38 | 1:120458122 | NOTCH2 | c.7223T>A | p.Leu2408His | Missense |
| 6:51914991 | PKHD1 | c.2243C>T | p.Ala748Val | Missense | |
| 16:23646942 | PALB2 | c.925A>G | p.Ile309Val | Missense | |
| 17:5462805 | NLRP1 | c.1211G>A | p.Arg404Gln | Missense | |
| 17:37864584 | ERBB2 | c.236A>C | p.Glu79Ala | Missense | |
| 3:65425588 | MAGI1 | c.1234_1236delCAG | p.Gln421del | Inframe - deletion | |
| 19:17937659 | JAK3 | c.3268G>A | p.Ala1090Thr | Missense | |
| ULMS39 | 3:188327501 | LPP | c.982C>T | p.Arg328Trp | Missense |
| 7:142562071 | EPHB6 | c.513_515delCTC | p.Ser176del | LOF: disruptive - inframe - del | |
| 17:7577545 | TP53 | c.736A>G | p.Met246Val | Missense | |
| ULMS40 | 2:100218031 | AFF3 | c.1310_1312delGCA | p.Ser444del | LOF: disruptive - inframe - del |
| ULMS40 | 11:108139268 | ATM | c.2770C>T | p.Arg924Trp | Missense |
| 17:7577120 | TP53 | c.818G>A | p.Arg273His | Missense | |
| 17:37881117 | ERBB2 | c.2446C>T | p.Arg816Cys | Missense | |
| X:76891445 | ATRX | c.4660A>T | p.Arg1554Ter | LOF: stop - gained | |
| ULMS45 | 3:128204775 | GATA2 | c.666G>C | p.Lys222Asn | Missense |
| 11:108160506 | ATM | c.4414T>G | p.Leu1472Val | Missense | |
| 12:121437187 | HNF1A | c.1618A>G | p.Lys540Glu | Missense | |
| 17:7578290 | TP53 | c.560-1G>C | r.spl? | LOF: splice - acceptor | |
| 16:2135281 | TSC2 | c.4620C>A | p.Tyr1540Ter | LOF: stop - gained | |
| ULMS50b | 2:29432740 | ALK | c.3748A>G | p.Ile1250Val | Missense |
| ULMS52 | 1:6528318 | PLEKHG5 | c.2815C>T | p.Arg939Cys | Missense |
| 1:120459251 | NOTCH2 | c.6094C>A | p.His2032Asn | Missense | |
| 9:139400980 | NOTCH1 | c.4013C>T | p.Ala1338Val | Missense | |
| 11:118377142 | KMT2A | c.10535C>T | p.Pro3512Leu | Missense | |
| 12:49416396 | KMT2D | c.16315C>T | p.Arg5439Trp | Missense | |
| 13:26978093 | CDK8 | c.1270C>T | p.Arg424Cys | Missense | |
| 16:2130319 | TSC2 | c.3551C>T | p.Ala1184Val | Missense | |
| 16:3779521 | CREBBP | c.5527T>C | p.Cys1843Arg | Missense | |
| 16:3790470 | CREBBP | c.4063G>A | p.Gly1355Arg | Missense | |
| 17:7574017 | TP53 | c.1010G>A | p.Arg337His | Missense | |
| 22:36678790 | MYH9 | c.5807G>A | p.Arg1936Gln | Missense | |
| ULMS59 | 5:112173857 | APC | c.2566C>T | p.Arg856Cys | Missense |
| 6:135511289 | MYB | c.331G>A | p.Gly111Ser | Missense | |
| 6:135539101 | MYB | c.2269C>T | p.Arg757Trp | Missense | |
| 6:152832196 | SYNE1 | c.352C>T | p.Arg118Ter | LOF: stop - gained | |
| 7:2946463 | CARD11 | c.3274C>T | p.Arg1092Ter | LOF: stop - gained | |
| 18:22806393 | ZNF521 | c.1489C>T | p.Arg497Ter | LOF: stop - gained | |
| 18:47803035 | MBD1 | c.472C>T | p.Arg158Ter | LOF: stop - gained | |
| 20:57429026 | GNAS | c.706G>A | p.Asp236Asn | Missense | |
| 20:57480457 | GNAS | c.2381A>C | p.Lys794Thr | Missense | |
| 22:30069262 | NF2 | c.1127G>A | p.Arg376Gln | Missense | |
| ESS2 (LG-ESS) | 6:152706896 | SYNE1 | c.8565G>A | p.Trp2855Ter | LOF: stop - gained |
| 11:108175463 | ATM | c.5558A>T | p.Asp1853Val | Missense | |
| 14:81610269 | TSHR | c.1867G>T | p.Ala623Ser | Missense | |
| 17:7577121 | TP53 | c.817C>T | p.Arg273Cys | Missense | |
| 17:7577139 | TP53 | c.799C>T | p.Arg267Trp | Missense | |
| 19:3119273 | GNA11 | c.805G>A | p.Val269Ile | Missense | |
| 22:41553308 | EP300 | c.3397C>T | p.Arg1133Trp | Missense | |
| ESS3 | 1:145015874 | PDE4DIP | c.214C>T | p.Arg72Ter | LOF: stop - gained |
| 5:112154777 | APC | c.1048T>C | p.Ser350Pro | Missense | |
| 5:112162855 | APC | c.1459G>A | p.Gly487Arg | Missense | |
| 6:56328464 | DST | c.16429C>T | p.Arg5477Trp | Missense | |
| 12:49418436 | KMT2D | c.15977T>C | p.Leu5326Pro | Missense | |
| 17:7578176 | TP53 | c.672+1G>A | r.spl? | LOF: splice - donor | |
| 17:29556250 | NF1 | c.2617C>T | p.Arg873Cys | Missense | |
| 17:29677234 | NF1 | c.7355G>T | p.Arg2452Leu | Missense | |
| ESS4 | 11:108141990 | ATM | c.2934delT | p.Leu979Cysfs | LOF: frameshift |
| 16:3820773 | CREBBP | c.2678C>T | p.Ser893Leu | Missense | |
| ESS5 | 1:11217330 | MTOR | c.4348T>G | p.Tyr1450Asp | Missense |
| 14:51227050 | NIN | c.1924G>A | p.Glu642Lys | Missense | |
| 19:17937659 | JAK3 | c.3268G>A | p.Ala1090Thr | Missense | |
| 20:41101170 | PTPRT | c.1186G>A | p.Val396Ile | Missense | |
| ESS7 | 6:33287248 | DAXX | c.1885G>A | p.Val629Ile | Missense |
| 6:117710646 | ROS1 | c.1626delT | p.Phe542Leufs | LOF: frameshift | |
| 14:95590677 | DICER1 | c.1232C>A | p.Ser411Ter | LOF: stop - gained | |
| ESS7 | X:76939115 | ATRX | c.1633C>G | p.Gln545Glu | Missense |
| ESS9 | 1:144906139 | PDE4DIP | c.2494delC | p.Gln832Argfs | LOF: frameshift |
| 1:145536012 | ITGA10 | c.2104G>A | p.Ala702Thr | Missense | |
| 3:178936091 | PIK3CA | c.1633G>A | p.Glu545Lys | Missense | |
| 4:55564641 | KIT | c.529C>T | p.Arg177Cys | Missense | |
| 4:55976709 | KDR | c.1116G>C | p.Glu372Asp | Missense | |
| 5:112175711 | APC | c.4420G>A | p.Ala1474Thr | Missense | |
| 5:180048651 | FLT4 | c.1911C>G | p.Ser637Arg | Missense | |
| ESS58107 | 1:145015874 | PDE4DIP | c.214C>T | p.Arg72Ter | LOF: stop - gained |
| 1:145536012 | ITGA10 | c.2104G>A | p.Ala702Thr | Missense | |
| 2:142567932 | LRP1B | c.121G>A | p.Asp41Asn | Missense | |
| 4:153332477 | FBXW7 | c.479C>T | p.Pro160Leu | Missense | |
| 6:56328464 | DST | c.16429C>T | p.Arg5477Trp | Missense | |
| 6:135516944 | MYB | c.1007C>T | p.Thr336Ile | Missense | |
| 7:91570414 | AKAP9 | c.1A>G | p.Met1? | LOF: initiator - codon | |
| 17:7578176 | TP53 | c.672+1G>A | r.spl? | LOF: splice - donor | |
| X:41056743 | USP9X | c.4360delG | p.Gly1454Glufs | LOF: frameshift | |
| X:66863156 | AR | c.1675A>T | p.Thr559Ser | Missense | |
| ADS2 | 1:162748436 | DDR2 | c.2350T>C | p.Cys784Arg | Missense |
| 2:25467477 | DNMT3A | c.1599C>A | p.Tyr533Ter | LOF: stop - gained | |
| 2:209110123 | IDH1 | c.440C>A | p.Pro147His | Missense | |
| 3:38182306 | MYD88 | c.766T>C | p.Phe256Leu | Missense | |
| 5:131927073 | RAD50 | c.1610delA | p.Met538Trpfs | LOF: frameshift | |
| 6:33288629 | DAXX | c.959A>G | p.Gln320Arg | Missense | |
| 6:93979315 | EPHA7 | c.1513C>A | p.Leu505Met | Missense | |
| 7:98501128 | TRRAP | c.1024G>T | p.Glu342Ter | LOF: stop - gained | |
| 8:48711786 | PRKDC | c.10279G>T | p.Glu3427Ter | LOF: stop - gained | |
| 9:98209391 | PTCH1 | c.4147C>A | p.Pro1383Thr | Missense | |
| 10:76781925 | KAT6B | c.3308_3310delAAG | p.Glu1104del | LOF: disruptive - inframe - del | |
| 11:106558436 | GUCY1A2 | c.2131G>T | p.Glu711Ter | LOF: stop - gained | |
| 15:90630454 | IDH2 | c.857A>G | p.Glu286Gly | Missense | |
| 16:2138078 | TSC2 | c.5098G>T | p.Ala1700Ser | Missense | |
| 22:29121048 | CHEK2 | c.638T>C | p.Val213Ala | Missense | |
| X:53223847 | KDM5C | c.3512A>G | p.Lys1171Arg | Missense | |
| ULM119 | 7:116403114 | MET | c.2429A>C | p.His810Pro | Missense |
| 22:39621795 | PDGFB | c.659dupA | p.Lys222Glnfs | LOF: frameshift | |
| ULM143 | 1:11307996 | MTOR | c.995_996dupGG | p.Leu333Glyfs | LOF: frameshift |
| 9:32634260 | TAF1L | c.1318A>G | p.Ile440Val | Missense | |
| 19:17945696 | JAK3 | c.2164G>A | p.Val722Ile | Missense | |
| ULM152 | 8:41791030 | KAT6A | c.4708G>A | p.Asp1570Asn | Missense |
| 11:118344893 | KMT2A | c.3019G>T | p.Gly1007Cys | Missense | |
| 19:1207176 | STK11 | c.263_264insC | p.Asn90Glnfs | LOF: frameshift |
Based on the data described in Table 4, we selected genes with more than three mutations in our cohort to submit to the OncoPrinter visualization tool (cBioPortal - http://www.cbioportal.org/). Figure 2 shows the percentage of patients demonstrating mutations in each gene, distribution, and the types of mutations observed in each sample. The highest frequency of gene mutations was observed in TP53 (48%) with the highest frequency of missense-type mutations (3 ULMS, 1 ESS, and 4 UCS samples). ATM mutations were observed in 22% of the samples, with 3 missense-type mutations (2 ULMS and 1 ESS) and 2 LOF-type mutations (1 ESS and 1 UCS). PIK3CA appeared to be the third most mutated gene (17%) present in 3 UCS samples, with most of the mutations determined as the missense-type. APC, MTOR, DICER1, TRRAP, KMT2D, TSC2, PDE4DIP, and JAK3 showed a 13% mutational frequency. LOF mutations in PDE4DIP was found exclusively/specificaly in the ESS samples. NF1, CREBBP, and MYB demonstrated a 9% mutational frequency. Missense mutations in CREBBP and MYB were associated with ULMS and ESS (4 mutations in ULMS and 2 in ESS).
Distribution of mutations in samples and their biological effects. The figure was constructed using the OncoPrinter from cBioPortal for Cancer Genomics database (http://www.cbioportal.org/). Each gray rectangle represents a sample according to the sequence indicated at the top. Genes with the highest frequency of alterations are shown. Captions for each type of alteration (Loss of function - Black Square; Missense - Green Square; Two alterations in the same gene - vertical line [modified by authors]; No alteration - gray rectangle) are indicated.
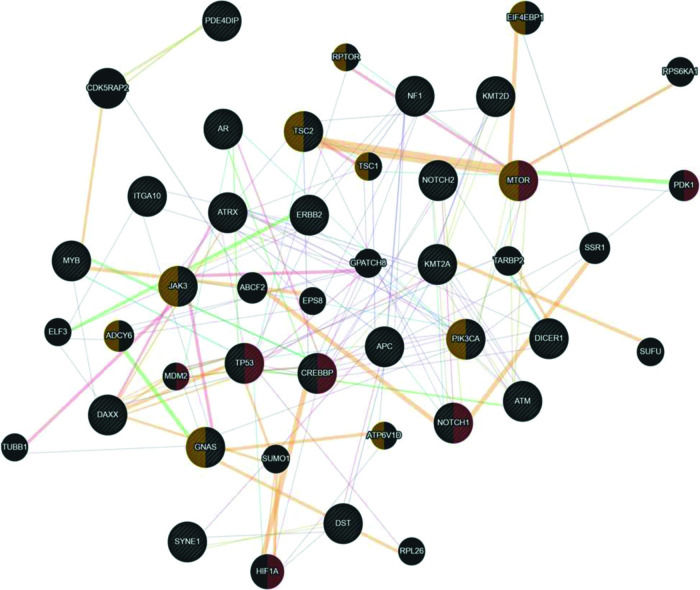
Since uterine sarcomas are histologically classified into two primary subtypes, we used the same classification to study the association of the mutated genes with pure sarcomas (ULMS - ESS) and mixed tumors (UCS - ADS). Figure 3 shows the association of the mutated genes in the group of tumors classified as pure (ULMS and ESS). According to the Cytoscape platform (20), many genes demonstrating mutations in these histological subtypes exhibit functions associated with the cellular response to hypoxia (MTOR, PDK1, MDM2, TP53, CREBBP, NOTCH1, and HIF1A) and peptide hormone stimulus (EIF4EBP1, RPTOR, TSC2, TSC1, MTOR, JAK3, ADCY6, PIK3CA, GNAS, and ATP6V1D).
Interaction network of mutated genes in the histological types of pure sarcomas (ULMS - ESS) prepared by the Cytoscape 3.7.0 platform. The network shows patterns of predicted interaction (orange); physical interactions (red); co-expression (violet); shared proteins domains (yellow); co-localization (blue), and genetic interaction (green). Red-labeled genes have a function associated with the cellular response to hypoxia and yellow-labeled genes have a function associated with the cellular response to the peptide hormone stimulus. The genes that were inserted to perform the analysis are shown with cross-hatched circles of a uniform size. The relevant genes are shown with solid circles whose size is proportional to the number of interactions. The reported link weights are indicated visually by line thickness.
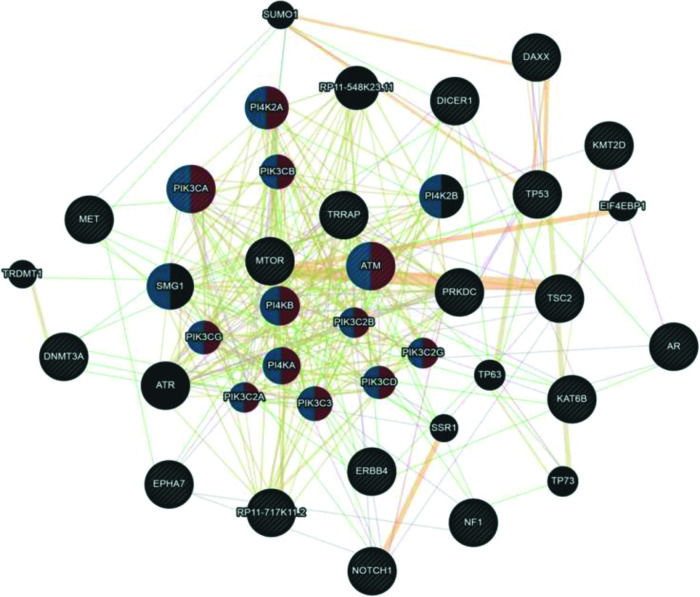
Although UCS is no longer classified as uterine sarcoma but as metaplastic carcinoma, we included this tumor group in the analysis shown in Figure 4. Here, we associated UCS - ADS owing to their mixed histologies (epithelial and mesenchymal components) and also because many retrospective studies on the US still include UCS in their available samples. According to the Cytoscape platform (20), many mutated genes in these tumors have functions associated with phosphatidylinositol kinase activity (PI4K2A, PIK3CA, PIK3CB, ATM, PI4KB, PIK3CG, PIK3C2B, PI4KA, PIK3C2A, PIK3C3, PIK3C2G, and PIK3CD) and glycerophospholipid metabolic process (PI4K2A, PIK3CA, PIK3CB, ATM, PI4KB, PIK3CG, PIK3C2B, PI4KA, PIK3C2A, PIK3C3, PIK3C2G, PIK3CD, PI4K2B, and SMG1).
Interaction network of mutated genes in mixed tumors (UCS - ADS) prepared by the Cytoscape 3.7.0 platform. The network shows patterns of predicted interaction (orange); physical interactions (red); co-expression (violet); shared proteins domains (yellow); co-localization (blue) and genetic interaction (green). Red-labeled genes have a function associated with phosphatidylinositol kinase activity and blue-labeled genes have a function associated with the glycerophospholipid metabolic process. The genes that were inserted to perform the analysis are shown with cross-hatched circles of a uniform size. The relevant genes are shown with solid circles whose size is proportional to the number of interactions. The reported link weights are indicated visually by line thickness.
Collectively, our results indicate that despite the molecular heterogeneity demonstrated by USs and UCSs, they share similarities in their mutational profiles. In addition, genetic interaction networks indicate that alterations in functions associated with hypoxia, response to peptide hormone stimulus in ULMSs and ESSs, and phosphatidylinositol kinase activity and glycerophospholipid metabolic process in UCS and ADS can influence the carcinogenic process of these tumors. Considering that NGS technology can provide a reliable molecular portrait of neoplasms quickly and cost-effectively (21), these results open new avenues for research and consequently, may positively impact the clinical management of patients with such tumors.
DISCUSSIONIn this study, we performed a mutational screening of the samples collected from patients with USs and UCSs. We employed a panel of 409 genes for the screening. Initially, we focused on the mutated genes shared among more than one histological subtype of US. We initiated our analyses with 40 samples, but owing to the quality of the FFPE material, certain losses reduced the number of samples to 23. Considering the published reports on sarcomas, the number of samples was sufficient for this type of population mutational screening. In UCS and ESS samples, we identified mutations in genes that demonstrated alterations in previous studies conducted for examining other tumors, such as PIK3CA, DICER1, AR, and NF (22). Although the role of these genes is known in different cancers, their role in the tumorigenesis of USs and USCs is not fully understood.
The PIK3CA gene encodes the p110α protein, the catalytic subunit of PI3K, which controls the growth, division, survival, movement, and structure of cells. Many studies have demonstrated the importance of PIK3CA mutation in mediating tumorigenesis via increased PI3K/AKT/mTOR signaling (23,24). While investigating druggable molecular targets in uterine sarcomas, Cuppens et. al (25) identified PI3K/MTOR as a potential target in 26% of cases, which were primarily ULMS, HG-ESS, and undifferentiated uterine sarcomas. Here, we included eight samples of ESS. Seven of these were characterized as HG-ESS, consistent with the molecular findings described in previous reports published for these tumors. DICER1 is critical for the regulation of expression of several miRNAs. The DICER1 gene is highly conserved among various species, indicating that mutations may compromise its function and might be involved in the onset of tumors (26). Previous reports published by our group (2,27) demonstrated the regulation of microRNAs associated with several oncogenic pathways, including DICER1. Mutations in NF1 have already been demonstrated in soft-tissue sarcomas (myxofibrosarcomas and pleomorphic liposarcomas) (28). The expression of the androgen receptor (AR) seems to be associated with a better prognosis in patients with ESS. AR expression is higher in pre-malignant lesions and low-grade tumors (LG-ESS) (29). These findings may explain why AR expression is low in ULMS, which is an extremely aggressive tumor (30). However, the effects of the mutations observed in this gene need to be further investigated for US.
It is important to note that NOTCH1 was the unique gene that shared mutations in the UCS and ULMS. Similarly, mutations in the DAXX gene have also been observed in the cases of ESS/ADS and ULMS/ADS, which share mutations in TSC2. Thus, our results suggest that besides exhibiting a similar tumor microenvironment, USs and UCSs also share genetic alterations. This observation is relevant to the understanding of the onset and evolution of these tumors. Furthermore, ULMS cases originating from ULMs have been reported; however, this hypothesis has not been proven yet (31,32). Our study showed that mutations in KMT2A were exclusively observed in ULMS and ULM. The c.3019G>T variant appears to be related to the Wiedemann-Steiner syndrome and Kabuki syndrome (33,34).
We attempted to identify specific genes for each type of tumor, establishing individual signatures. Despite the heterogeneity, we were able to identify six specific genes for three of the histological types evaluated in this study. In ESS samples, we observed variants in the PDE4DIP (c.214C>T and c.2494delC), ITGA10 (c.2104G>A), and DST (c.16429C>T) genes. The variant PDE4DIP c.214C>T is described in the COSMIC database (35) and was first observed in papillary thyroid carcinoma. Mutations in this gene are described in several tumors, such as breast cancer as well as the cancers of the endometrium, cervix, ovaries, and urinary tract. The protein encoded by the PDE4DIP gene is responsible for binding 4D phosphodiesterase to the Golgi complex. Alterations in this gene may cause a myeloproliferative disorder associated with eosinophilia (36). Despite the information available in databases and the literature, its typical role in tumor biology remains unknown.
In UCS, we observed two variants of ERBB4 (c.782A>C and c.2513G>A). The variant ERBB4 c.2513G>A is described in the COSMIC database (35) as pathogenic (score 0.99) and has already been observed in hormone receptor-positive breast cancer, large bowel adenocarcinoma, malignant melanoma, and gastroesophageal junction adenocarcinoma. The role of ERBB4 as a tumor progression factor is not fully elucidated. However, this gene is known to be overexpressed and/or mutated in several solid tumors (37). The monoclonal antibody ERBB4 therapy is effective in breast, lung, and prostate cancer cells in vitro and in vivo (38). Specific and detailed studies may demonstrate new opportunities for the development of therapies targeting these tumors.
Mutations in NOTCH2 and HER2 have also been observed exclusively in ULMS. All variants are described in the COSMIC database (35). c.6094C>A mutation of NOTCH2 is considered to be pathogenic (score 0.97) and is described in diffuse large B cell lymphoma and pancreatic ductal adenocarcinoma (PDAC). The NOTCH2 c.7223T>A variant is also pathogenic (score 0.85) and has already been described in meningioma, a primary non-malignant CNS tumor (39). HER2 also presented two pathogenic variants in ULMS: c.236A>C and c.2446C>T. The c.236A>C variant has already been described in meningothelial meningioma and is associated with IL-6 signaling pathways and DNA damage response. The c.2446C>T mutation has been observed in large bowel adenocarcinoma and transitional cell carcinoma of the urinary system. Persistent NOTCH2 signaling is largely associated with poor clinical prognosis. In addition, it increases resistance to chemotherapy and radiotherapy, making these cancers less sensitive to treatment (40). HER2 mutations have emerged as therapeutic targets for a variety of tumors. Anti- HER2 therapies are effective against breast, lung, and cervical cancers (41).
In this study, we were able to identify several mutations that contribute to a better understanding of the biology of USs and UCSs. Even with the limitations associated with rare tumors, we identified genetic alterations that might act as potential target markers for precision medicine-based approaches upon validation in larger cohorts. To date, there is no precise preoperative diagnostic test for these tumors. Although rare, such tumors are very aggressive and associated with a poor prognosis. Thus, even with small cohorts, the molecular profiling of USs and UCSs is extremely important to identify the changes driving the development of these tumors and provide powerful tools for diagnostic and prognostic tests as well as adequate treatment alternatives. Our study is the first DNA-sequencing study to investigate all histological types of USs and UCSs together and is an insightful contribution for defining the mutational repertoire of these rare tumors.
CONCLUSIONSUsing a platform to profile mutations in a panel of 409 genes, we identified that TP53, ATM, PIK3CA, APC, MTOR, DICER1, TRRAP, KMT2D, TSC2, PDE4DIP, and JAK3 are the most frequently mutated genes in USs and UCSs. Considering common mutations among the different tumor types being evaluated, the TP53 (4 UCS/4 ULMS/3 ESS), ATM (2 ULMS/2 ESS/1 UCS), and KMT2D (1 UCS/1 ULMS/1 ESS) genes could be indicators of similarities in neoplastic progression. As specific signature genes, ESS exhibited mutations in the PDE4DIP, IGTA10, and DST genes. UCS showed mutations in the ERBB4 gene, and ULMS demonstrated exclusive alterations in the NOTCH2 and HER2 genes. Mutations in the KMT2A gene were observed exclusively in ULM and ULMS samples, and therefore, are potentially involved in the malignant transformation process. According to the Cytoscape platform, many genes that were mutated in the ULMS and ESS samples exhibit functions associated with the cellular response to hypoxia and peptide hormone stimulus. In UCS and ADS, most altered genes exhibit functions associated with phosphatidylinositol kinase activity and glycerophospholipid metabolic process. More studies should be conducted with a larger number of samples and functional analyses. However, the current screening contributes to the characterization of the complex genetic profile of USs and USCs.
AUTHOR CONTRIBUTIONSDa Costa LT and Dos Anjos LG were responsible for study conceptualization, literature organization and paper elaboration. Kagohara LT collaborated in analyses of data, manuscripts and reviews. Torrezan GT and De Paula CAA contributed to the study execution. Baracat EC and Carraro DM provided intellectual support. Carvalho KC analyzed the literature, critically reviewed the manuscript, supervised the research and developed the original idea.
The research received financial support from Fundacão de Amparo è Pesquisa do Estado de São Paulo - FAPESP (process numbers: 2016/03163-6 and 2019/01109-2).
Ion AmpliSeq Comprehensive Cancer Panel target gene list.
| 1 | ABL1 | CBL | EP300 | GATA2 | LAMP1 | MYD88 | PKHD1 | SMARCA4 | WHSC1 |
| 2 | ABL2 | CCND1 | EP400 | GATA3 | LCK | MYH11 | PLAG1 | SMARCB1 | WRN |
| 3 | ACVR2A | CCND2 | EPHA3 | GDNF | LIFR | MYH9 | PLCG1 | SMO | WT1 |
| 4 | ADAMTS20 | CCNE1 | EPHA7 | GNA11 | LPHN3 | NBN | PLEKHG5 | SMUG1 | XPA |
| 5 | AFF1 | CD79A | EPHB1 | GNAQ | POT1 | NCOA1 | PML | SOCS1 | XPC |
| 6 | AFF3 | CD79B | EPHB4 | GNAS | LPP | NCOA2 | PMS1 | SOX11 | XPO1 |
| 7 | AKAP9 | CDC73 | EPHB6 | GPR124 | LRP1B | NCOA4 | PMS2 | SOX2 | XRCC2 |
| 8 | AKT1 | CDH1 | ERBB2 | GRM8 | LTF | NF1 | POU5F1 | SRC | ZNF384 |
| 9 | AKT2 | CDH11 | ERBB3 | GUCY1A2 | LTK | NF2 | PPARG | SSX1 | ZNF521 |
| 10 | AKT3 | CDH2 | ERBB4 | HCAR1 | MAF | NFE2L2 | PPP2R1A | STK11 | |
| 11 | ALK | CDH20 | ERCC1 | HIF1A | MAFB | NFKB1 | PRDM1 | STK36 | |
| 12 | APC | CDH5 | ERCC2 | HLF | MAGEA1 | NFKB2 | PRKAR1A | SUFU | |
| 13 | AR | CDK12 | ERCC3 | HNF1A | MAGI1 | NIN | PRKDC | SYK | |
| 14 | ARID1A | CDK4 | ERCC4 | HOOK3 | MALT1 | NKX2-1 | PSIP1 | SYNE1 | |
| 15 | ARID2 | CDK6 | ERCC5 | HRAS | MAML2 | NLRP1 | PTCH1 | TAF1 | |
| 16 | ARNT | CDK8 | ERG | HSP90AA1 | MAP2K1 | NOTCH1 | PTEN | TAF1L | |
| 17 | ASXL1 | CDKN2A | ESR1 | HSP90AB1 | MAP2K2 | NOTCH2 | PTGS2 | TAL1 | |
| 18 | ATF1 | CDKN2B | ETS1 | ICK | MAP2K4 | NOTCH4 | PTPN11 | TBX22 | |
| 19 | ATM | CDKN2C | ETV1 | IDH1 | MAP3K7 | NPM1 | PTPRD | TCF12 | |
| 20 | ATR | CEBPA | ETV4 | IDH2 | MAPK1 | NRAS | PTPRT | TCF3 | |
| 21 | ATRX | CHEK1 | EXT1 | IGF1R | MAPK8 | NSD1 | RAD50 | TCF7L1 | |
| 22 | AURKA | CHEK2 | EXT2 | IGF2 | MARK1 | NTRK1 | RAF1 | TCF7L2 | |
| 23 | AURKB | CIC | EZH2 | IGF2R | MARK4 | NTRK3 | RALGDS | TCL1A | |
| 24 | AURKC | CKS1B | FAM123B | IKBKB | MBD1 | NUMA1 | RARA | TET1 | |
| 25 | AXL | CMPK1 | FANCA | IKBKE | MCL1 | NUP214 | RB1 | TET2 | |
| 26 | BAI3 | COL1A1 | FANCC | IKZF1 | MDM2 | NUP98 | RECQL4 | TFE3 | |
| 27 | BAP1 | CRBN | FANCD2 | IL2 | MDM4 | PAK3 | REL | TGFBR2 | |
| 28 | BCL10 | CREB1 | FANCF | IL21R | MEN1 | PALB2 | RET | TGM7 | |
| 29 | BCL11A | CREBBP | FANCG | IL6ST | MET | PARP1 | RHOH | THBS1 | |
| 30 | BCL11B | CRKL | FAS | IL7R | MITF | PAX3 | RNASEL | TIMP3 | |
| 31 | BCL2 | CRTC1 | FBXW7 | ING4 | MLH1 | PAX5 | RNF2 | TLR4 | |
| 32 | BCL2L1 | CSF1R | FGFR1 | IRF4 | MLL | PAX7 | RNF213 | TLX1 | |
| 33 | BCL2L2 | CSMD3 | FGFR2 | IRS2 | MLL2 | PAX8 | ROS1 | TNFAIP3 | |
| 34 | BCL3 | CTNNA1 | FGFR3 | ITGA10 | MLL3 | PBRM1 | RPS6KA2 | TNFRSF14 | |
| 35 | BCL6 | CTNNB1 | FGFR4 | ITGA9 | MLLT10 | PBX1 | RRM1 | TNK2 | |
| 36 | BCL9 | CYLD | FH | ITGB2 | MMP2 | PDE4DIP | RUNX1 | TOP1 | |
| 37 | BCR | CYP2C19 | FLCN | ITGB3 | MN1 | PDGFB | RUNX1T1 | TP53 | |
| 38 | BIRC2 | CYP2D6 | FLI1 | JAK1 | MPL | PDGFRA | SAMD9 | TPR | |
| 39 | BIRC3 | DAXX | FLT1 | JAK2 | MRE11A | PDGFRB | SBDS | TRIM24 | |
| 40 | BIRC5 | DCC | FLT3 | JAK3 | MSH2 | PER1 | SDHA | TRIM33 | |
| 41 | BLM | DDB2 | FLT4 | JUN | MSH6 | PGAP3 | SDHB | TRIP11 | |
| 42 | BLNK | DDIT3 | FN1 | KAT6A | MTOR | PHOX2B | SDHC | TRRAP | |
| 43 | BMPR1A | DDR2 | FOXL2 | KAT6B | MTR | PIK3C2B | SDHD | TSC1 | |
| 44 | BRAF | DEK | FOXO1 | KDM5C | MTRR | PIK3CA | SEPT9 | TSC2 | |
| 45 | BRD3 | DICER1 | FOXO3 | KDM6A | MUC1 | PIK3CB | SETD2 | TSHR | |
| 46 | BRIP1 | DNMT3A | FOXP1 | KDR | MUTYH | PIK3CD | SF3B1 | UBR5 | |
| 47 | BTK | DPYD | FOXP4 | KEAP1 | MYB | PIK3CG | SGK1 | UGT1A1 | |
| 48 | BUB1B | DST | FZR1 | KIT | MYC | PIK3R1 | SH2D1A | USP9X | |
| 49 | CARD11 | EGFR | G6PD | KLF6 | MYCL1 | PIK3R2 | SMAD2 | VHL | |
| 50 | CASC5 | EML4 | GATA1 | KRAS | MYCN | PIM1 | SMAD4 | WAS |
Description of 153 potential somatic variants selected in 23 samples of uterine tumors.
| Sample | Chr:Pos | Gene | HGVS c. | HGVS p. | Effect |
|---|---|---|---|---|---|
| UCS2 | 3:178921549 | PIK3CA | c.1031T>C | p.Val344Ala | Missense |
| 6:94120318 | EPHA7 | c.733G>A | p.Ala245Thr | Missense | |
| 7:116339356 | MET | c.218T>A | p.Leu73Ter | LOF: stop - gained | |
| 8:48776121 | PRKDC | c.5586delT | p.Phe1862Leufs | LOF: frameshift | |
| 15:99500303 | IGF1R | c.3736C>T | p.Arg1246Cys | Missense | |
| 17:7577547 | TP53 | c.734G>T | p.Gly245Val | Missense | |
| 22:33253291 | TIMP3 | c.260delC | p.Glu88Argfs | LOF: frameshift | |
| UCS5 | 1:11227575 | MTOR | c.4254-1G>A | r.spl? | LOF: splice - acceptor |
| 1:27105553 | ARID1A | c.5164C>T | p.Arg1722Ter | LOF: stop - gained | |
| 1:65310574 | JAK1 | c.2116-2A>G | r.spl? | LOF: splice - acceptor | |
| 3:178952085 | PIK3CA | c.3140A>G | p.His1047Arg | Missense | |
| 10:76735809 | KAT6B | c.1714C>T | p.Arg572Cys | Missense | |
| 10:97969609 | BLNK | c.731C>T | p.Pro244Leu | Missense | |
| 11:108114777 | ATM | c.594C>A | p.Cys198Ter | LOF: stop - gained | |
| 14:95572101 | DICER1 | c.3007C>T | p.Arg1003Ter | LOF: stop - gained | |
| 17:29588751 | NF1 | c.4600C>T | p.Arg1534Ter | LOF: stop - gained | |
| 17:29665110 | NF1 | c.6772C>T | p.Arg2258Ter | LOF: stop - gained | |
| 19:45260400 | BCL3 | c.646C>T | p.Arg216Cys | Missense | |
| 1:47685756 | TAL1 | c.632G>A | p.Arg211His | Missense | |
| 2:25469168 | DNMT3A | c.1290T>G | p.Asn430Lys | Missense | |
| 2:212587219 | ERBB4 | c.782A>C | p.Gln261Pro | Missense | |
| 7:98513427 | TRRAP | c.2281C>T | p.Arg761Trp | Missense | |
| 9:37015073 | PAX5 | c.331G>A | p.Ala111Thr | Missense | |
| 19:11098401 | SMARCA4 | c.919C>T | p.Pro307Ser | Missense | |
| 20:36030940 | SRC | c.1219G>A | p.Asp407Asn | Missense | |
| X:44942716 | KDM6A | c.3452A>G | p.Gln1151Arg | Missense | |
| X:66766207 | AR | c.1219C>T | p.Arg407Cys | Missense | |
| UCS9 | 9:139391355 | NOTCH1 | c.6836C>T | p.Ala2279Val | Missense |
| 10:123298226 | FGFR2 | c.628C>T | p.Arg210Ter | LOF: stop - gained | |
| 12:49444719 | KMT2D | c.2747C>T | p.Pro916Leu | Missense | |
| 15:40916649 | KNL1 | c.4265G>A | p.Arg1422Gln | Missense | |
| 17:7578442 | TP53 | c.488A>G | p.Tyr163Cys | Missense | |
| 3:52440867 | BAP1 | c.637C>T | p.Arg213Cys | Missense | |
| 21:39755729 | ERG | c.1057G>A | p.Glu353Lys | Missense | |
| UCS13 | 3:178916854 | PIK3CA | c.241G>A | p.Glu81Lys | Missense |
| 11:71726283 | NUMA1 | c.2266G>T | p.Glu756Ter | LOF: stop - gained | |
| 13:29001422 | FLT1 | c.1310C>T | p.Ser437Leu | Missense | |
| 14:95574253 | DICER1 | c.2614G>A | p.Ala872Thr | Missense | |
| 17:7577534 | TP53 | c.747G>T | p.Arg249Ser | Missense | |
| 5:176636902 | NSD1 | c.1502A>G | p.Lys501Arg | Missense | |
| 7:98609947 | TRRAP | c.11549G>A | p.Arg3850His | Missense | |
| UCS19 | 2:212295800 | ERBB4 | c.2513G>A | p.Arg838Gln | Missense |
| 9:5126715 | JAK2 | c.3323A>G | p.Asn1108Ser | Missense | |
| 17:7577580 | TP53 | c.701A>G | p.Tyr234Cys | Missense | |
| 17:37829120 | PGAP3 | c.900-1G>A | r.spl? | LOF: splice - acceptor | |
| ULMS38 | 1:120458122 | NOTCH2 | c.7223T>A | p.Leu2408His | Missense |
| 6:51914991 | PKHD1 | c.2243C>T | p.Ala748Val | Missense | |
| 16:23646942 | PALB2 | c.925A>G | p.Ile309Val | Missense | |
| 17:5462805 | NLRP1 | c.1211G>A | p.Arg404Gln | Missense | |
| 17:37864584 | ERBB2 | c.236A>C | p.Glu79Ala | Missense | |
| 3:65425588 | MAGI1 | c.1234_1236delCAG | p.Gln421del | Inframe - deletion | |
| 19:17937659 | JAK3 | c.3268G>A | p.Ala1090Thr | Missense | |
| ULMS39 | 3:188327501 | LPP | c.982C>T | p.Arg328Trp | Missense |
| 7:142562071 | EPHB6 | c.513_515delCTC | p.Ser176del | LOF: disruptive - inframe - del | |
| 17:7577545 | TP53 | c.736A>G | p.Met246Val | Missense | |
| ULMS40 | 2:100218031 | AFF3 | c.1310_1312delGCA | p.Ser444del | LOF: disruptive - inframe - del |
| ULMS40 | 11:108139268 | ATM | c.2770C>T | p.Arg924Trp | Missense |
| 17:7577120 | TP53 | c.818G>A | p.Arg273His | Missense | |
| 17:37881117 | ERBB2 | c.2446C>T | p.Arg816Cys | Missense | |
| X:76891445 | ATRX | c.4660A>T | p.Arg1554Ter | LOF: stop - gained | |
| ULMS45 | 3:128204775 | GATA2 | c.666G>C | p.Lys222Asn | Missense |
| 11:108160506 | ATM | c.4414T>G | p.Leu1472Val | Missense | |
| 12:121437187 | HNF1A | c.1618A>G | p.Lys540Glu | Missense | |
| 17:7578290 | TP53 | c.560-1G>C | r.spl? | LOF: splice - acceptor | |
| 16:2135281 | TSC2 | c.4620C>A | p.Tyr1540Ter | LOF: stop - gained | |
| ULMS50b | 2:29432740 | ALK | c.3748A>G | p.Ile1250Val | Missense |
| ULMS52 | 1:6528318 | PLEKHG5 | c.2815C>T | p.Arg939Cys | Missense |
| 1:120459251 | NOTCH2 | c.6094C>A | p.His2032Asn | Missense | |
| 9:139400980 | NOTCH1 | c.4013C>T | p.Ala1338Val | Missense | |
| 11:118377142 | KMT2A | c.10535C>T | p.Pro3512Leu | Missense | |
| 12:49416396 | KMT2D | c.16315C>T | p.Arg5439Trp | Missense | |
| 13:26978093 | CDK8 | c.1270C>T | p.Arg424Cys | Missense | |
| 16:2130319 | TSC2 | c.3551C>T | p.Ala1184Val | Missense | |
| 16:3779521 | CREBBP | c.5527T>C | p.Cys1843Arg | Missense | |
| 16:3790470 | CREBBP | c.4063G>A | p.Gly1355Arg | Missense | |
| 17:7574017 | TP53 | c.1010G>A | p.Arg337His | Missense | |
| 22:36678790 | MYH9 | c.5807G>A | p.Arg1936Gln | Missense | |
| ULMS59 | 5:112173857 | APC | c.2566C>T | p.Arg856Cys | Missense |
| 6:135511289 | MYB | c.331G>A | p.Gly111Ser | Missense | |
| 6:135539101 | MYB | c.2269C>T | p.Arg757Trp | Missense | |
| 6:152832196 | SYNE1 | c.352C>T | p.Arg118Ter | LOF: stop - gained | |
| 7:2946463 | CARD11 | c.3274C>T | p.Arg1092Ter | LOF: stop - gained | |
| 18:22806393 | ZNF521 | c.1489C>T | p.Arg497Ter | LOF: stop - gained | |
| 18:47803035 | MBD1 | c.472C>T | p.Arg158Ter | LOF: stop - gained | |
| 20:57429026 | GNAS | c.706G>A | p.Asp236Asn | Missense | |
| 20:57480457 | GNAS | c.2381A>C | p.Lys794Thr | Missense | |
| 22:30069262 | NF2 | c.1127G>A | p.Arg376Gln | Missense | |
| ESS2 (LG-ESS) | 6:152706896 | SYNE1 | c.8565G>A | p.Trp2855Ter | LOF: stop - gained |
| 11:108175463 | ATM | c.5558A>T | p.Asp1853Val | Missense | |
| 14:81610269 | TSHR | c.1867G>T | p.Ala623Ser | Missense | |
| 17:7577121 | TP53 | c.817C>T | p.Arg273Cys | Missense | |
| 17:7577139 | TP53 | c.799C>T | p.Arg267Trp | Missense | |
| 19:3119273 | GNA11 | c.805G>A | p.Val269Ile | Missense | |
| 22:41553308 | EP300 | c.3397C>T | p.Arg1133Trp | Missense | |
| ESS3 | 1:145015874 | PDE4DIP | c.214C>T | p.Arg72Ter | LOF: stop - gained |
| 5:112154777 | APC | c.1048T>C | p.Ser350Pro | Missense | |
| 5:112162855 | APC | c.1459G>A | p.Gly487Arg | Missense | |
| 6:56328464 | DST | c.16429C>T | p.Arg5477Trp | Missense | |
| 12:49418436 | KMT2D | c.15977T>C | p.Leu5326Pro | Missense | |
| 17:7578176 | TP53 | c.672+1G>A | r.spl? | LOF: splice - donor | |
| 17:29556250 | NF1 | c.2617C>T | p.Arg873Cys | Missense | |
| 17:29677234 | NF1 | c.7355G>T | p.Arg2452Leu | Missense | |
| ESS4 | 11:108141990 | ATM | c.2934delT | p.Leu979Cysfs | LOF: frameshift |
| 16:3820773 | CREBBP | c.2678C>T | p.Ser893Leu | Missense | |
| ESS5 | 1:11217330 | MTOR | c.4348T>G | p.Tyr1450Asp | Missense |
| 14:51227050 | NIN | c.1924G>A | p.Glu642Lys | Missense | |
| 19:17937659 | JAK3 | c.3268G>A | p.Ala1090Thr | Missense | |
| 20:41101170 | PTPRT | c.1186G>A | p.Val396Ile | Missense | |
| ESS7 | 6:33287248 | DAXX | c.1885G>A | p.Val629Ile | Missense |
| 6:117710646 | ROS1 | c.1626delT | p.Phe542Leufs | LOF: frameshift | |
| 14:95590677 | DICER1 | c.1232C>A | p.Ser411Ter | LOF: stop - gained | |
| ESS7 | X:76939115 | ATRX | c.1633C>G | p.Gln545Glu | Missense |
| ESS9 | 1:144906139 | PDE4DIP | c.2494delC | p.Gln832Argfs | LOF: frameshift |
| 1:145536012 | ITGA10 | c.2104G>A | p.Ala702Thr | Missense | |
| 3:178936091 | PIK3CA | c.1633G>A | p.Glu545Lys | Missense | |
| 4:55564641 | KIT | c.529C>T | p.Arg177Cys | Missense | |
| 4:55976709 | KDR | c.1116G>C | p.Glu372Asp | Missense | |
| 5:112175711 | APC | c.4420G>A | p.Ala1474Thr | Missense | |
| 5:180048651 | FLT4 | c.1911C>G | p.Ser637Arg | Missense | |
| ESS58107 | 1:145015874 | PDE4DIP | c.214C>T | p.Arg72Ter | LOF: stop - gained |
| 1:145536012 | ITGA10 | c.2104G>A | p.Ala702Thr | Missense | |
| 2:142567932 | LRP1B | c.121G>A | p.Asp41Asn | Missense | |
| 4:153332477 | FBXW7 | c.479C>T | p.Pro160Leu | Missense | |
| 6:56328464 | DST | c.16429C>T | p.Arg5477Trp | Missense | |
| 6:135516944 | MYB | c.1007C>T | p.Thr336Ile | Missense | |
| 7:91570414 | AKAP9 | c.1A>G | p.Met1? | LOF: initiator - codon | |
| 17:7578176 | TP53 | c.672+1G>A | r.spl? | LOF: splice - donor | |
| X:41056743 | USP9X | c.4360delG | p.Gly1454Glufs | LOF: frameshift | |
| X:66863156 | AR | c.1675A>T | p.Thr559Ser | Missense | |
| ADS2 | 1:162748436 | DDR2 | c.2350T>C | p.Cys784Arg | Missense |
| 2:25467477 | DNMT3A | c.1599C>A | p.Tyr533Ter | LOF: stop - gained | |
| 2:209110123 | IDH1 | c.440C>A | p.Pro147His | Missense | |
| 3:38182306 | MYD88 | c.766T>C | p.Phe256Leu | Missense | |
| 5:131927073 | RAD50 | c.1610delA | p.Met538Trpfs | LOF: frameshift | |
| 6:33288629 | DAXX | c.959A>G | p.Gln320Arg | Missense | |
| 6:93979315 | EPHA7 | c.1513C>A | p.Leu505Met | Missense | |
| 7:98501128 | TRRAP | c.1024G>T | p.Glu342Ter | LOF: stop - gained | |
| 8:48711786 | PRKDC | c.10279G>T | p.Glu3427Ter | LOF: stop - gained | |
| 9:98209391 | PTCH1 | c.4147C>A | p.Pro1383Thr | Missense | |
| 10:76781925 | KAT6B | c.3308_3310delAAG | p.Glu1104del | LOF: disruptive - inframe - del | |
| 11:106558436 | GUCY1A2 | c.2131G>T | p.Glu711Ter | LOF: stop - gained | |
| 15:90630454 | IDH2 | c.857A>G | p.Glu286Gly | Missense | |
| 16:2138078 | TSC2 | c.5098G>T | p.Ala1700Ser | Missense | |
| 22:29121048 | CHEK2 | c.638T>C | p.Val213Ala | Missense | |
| X:53223847 | KDM5C | c.3512A>G | p.Lys1171Arg | Missense | |
| ULM119 | 7:116403114 | MET | c.2429A>C | p.His810Pro | Missense |
| 22:39621795 | PDGFB | c.659dupA | p.Lys222Glnfs | LOF: frameshift | |
| ULM143 | 1:11307996 | MTOR | c.995_996dupGG | p.Leu333Glyfs | LOF: frameshift |
| 9:32634260 | TAF1L | c.1318A>G | p.Ile440Val | Missense | |
| 19:17945696 | JAK3 | c.2164G>A | p.Val722Ile | Missense | |
| ULM152 | 8:41791030 | KAT6A | c.4708G>A | p.Asp1570Asn | Missense |
| 11:118344893 | KMT2A | c.3019G>T | p.Gly1007Cys | Missense | |
| 19:1207176 | STK11 | c.263_264insC | p.Asn90Glnfs | LOF: frameshift |
No potential conflict of interest was reported.






![Distribution of mutations in samples and their biological effects. The figure was constructed using the OncoPrinter from cBioPortal for Cancer Genomics database (http://www.cbioportal.org/). Each gray rectangle represents a sample according to the sequence indicated at the top. Genes with the highest frequency of alterations are shown. Captions for each type of alteration (Loss of function - Black Square; Missense - Green Square; Two alterations in the same gene - vertical line [modified by authors]; No alteration - gray rectangle) are indicated. Distribution of mutations in samples and their biological effects. The figure was constructed using the OncoPrinter from cBioPortal for Cancer Genomics database (http://www.cbioportal.org/). Each gray rectangle represents a sample according to the sequence indicated at the top. Genes with the highest frequency of alterations are shown. Captions for each type of alteration (Loss of function - Black Square; Missense - Green Square; Two alterations in the same gene - vertical line [modified by authors]; No alteration - gray rectangle) are indicated.](https://static.elsevier.es/multimedia/18075932/000000760000000C/v1_202211191031/S1807593222000886/v1_202211191031/en/main.assets/thumbnail/gr2.jpeg?xkr=ue/ImdikoIMrsJoerZ+w96p5LBcBpyJTqfwgorxm+Ow=)

