Juvenile idiopathic arthritis (JIA) is an autoimmune rheumatic disease, which affects primarily the joints in children under 16 years old. The etiology of JIA is yet unknown but research has shown that JIA is a multifactorial disease implicating several genes and environmental factors. Environmental factors affect immune cells via epigenetic mechanisms. One of the most important epigenetic mechanisms is DNA methylation catalyzed by DNA methyltransferases (DNMTs) and usually associated with gene silencing. In this study, we analyzed the expression of three DNA methyltransferases namely DNMT1, DNMT3a and DNMT3b in peripheral blood mononuclear cells (PBMCs) of patients with JIA and compared it with the expression of these genes in healthy young individuals.
Materials and methodsPeripheral blood mononuclear cells of 28 JIA patients and 28 healthy controls were isolated. Total RNA was extracted, cDNA was synthesized and the transcript levels of DNMTs were analyzed by quantitative PCR.
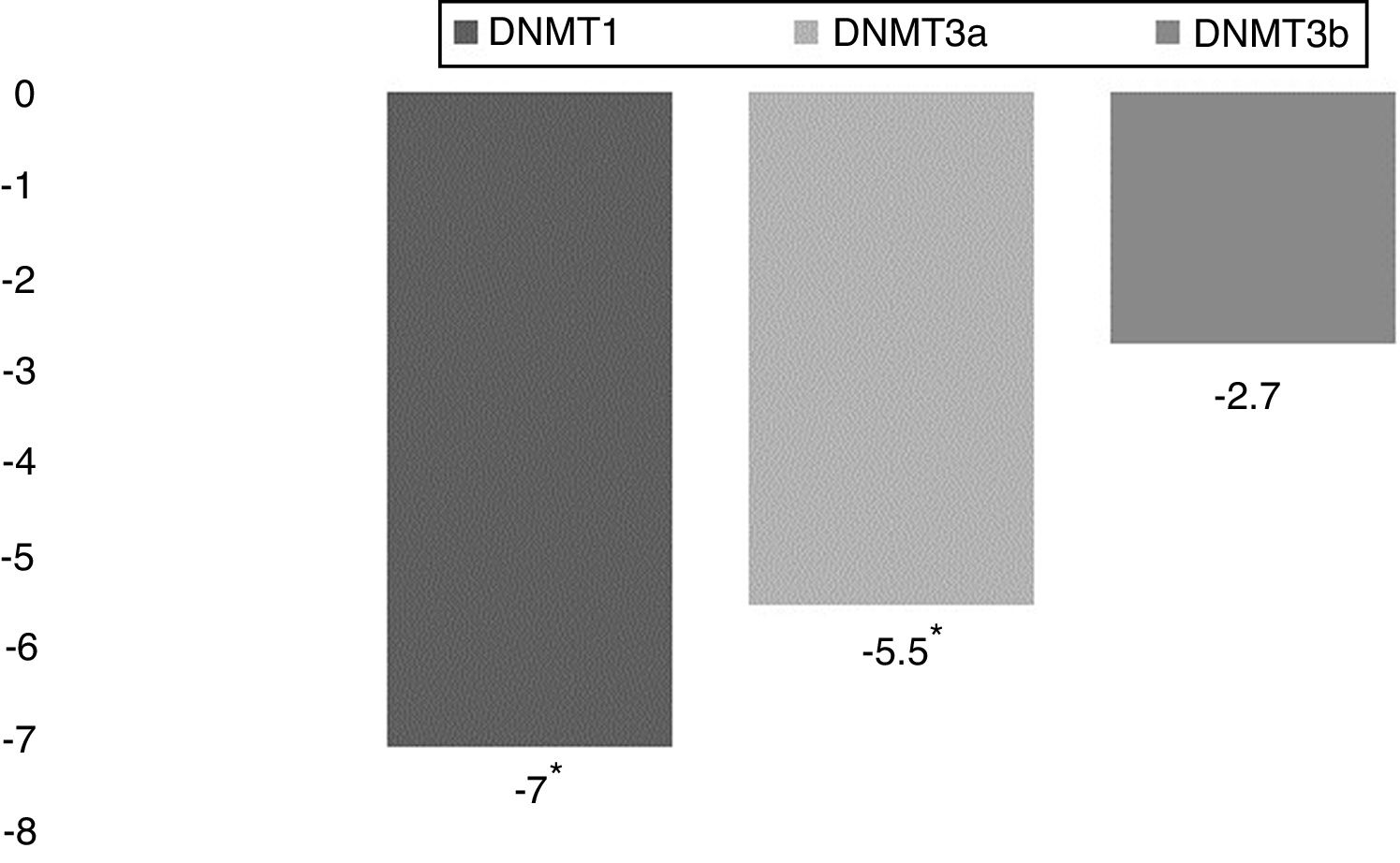
ResultsAnalysis of DNMT1, DNMT3a and DNMT3b relative gene expression in PBMCs of JIA patients and control individuals shows that the expression of DNMT1 and DNMT3a is reduced significantly by 7 folds and 5.5 folds, respectively, in JIA patients compared to healthy controls. Furthermore, the expression of all three DNMTs were significantly and drastically reduced in young affected males compared to healthy males.
ConclusionThis study shows that the expression of DNMTs is reduced in JIA patients and this reduction is severe in male JIA patients.
Juvenile Idiopathic Arthritis (JIA) is the most common autoimmune rheumatic disease characterized by the early onset of joint inflammation.1,2 According to the American College of Rheumatology (ACR), JIA is classified into three subtypes: oligoarticular, polyarticular and systemic. This classification is based on the number of affected joints, drug response and the special treatment needed by each group.3
The etiology of the disease is unknown but family studies4 suggest that JIA is a complex disease which means that genetic and environmental factors both have important roles.5 In this disease the immune system, specially T cells, attacks the body’s own tissues and causes damage.6
One way in which environmental factors affect the cell is via epigenetic changes. Epigenetics is the study of heritable changes in gene activity without effects on the DNA sequence. Different epigenetic mechanisms are used to change gene expression pattern. Histone modifications by histone methyltransferases (HMTs) and acetyltransferases )HATs) and DNA methylation by DNA methyltransferases (DNMTs) are part of these epigenetic mechanisms.7 It has been shown that some changes in epigenetic patterns induce several conditions including cancer and autoimmune disorders such as JIA.8
Three major types of DNMTs are identified in mammalian cells; DNMT1, which is required for the maintenance of DNA methylation during DNA replication, and DNMT3a and DNMT3b, which are de novo methyltransferases. However methylation of DNA during replication by DNMT3a and DNMT3b was also observed.9,10 It has been suggested that down regulation of DNMTs can decrease the methylation level of the genome and cause gene expression profile changes. Most studies show that environmental factors such as stress11 and cigarette smoke12 affect the expression of DNMTs. Here, we compared the expression of DNMT1, DNMT3a and DNMT3b in young patients affected with JIA with healthy young individuals.
We studied DNA methyl transferases’ (DNMTs) type 1, 3A and 3B transcript levels in peripheral blood mononuclear cells to confirm the importance of epigenetic alterations in JIA etiology. Previous study showed that in JIA CD4 + T cells, DNA methylation at the gene encoding the pro-inflammatory cytokine interleukin-32 (IL32) is reduced.13 According to this data, expression of unmethylated cytokine upregulates and causes hyperactivity of immune system, which could cause autoimmune disease. This growing evidence suggesting that reducing of DNA methylation is linked to the pathogenesis of JIA therefore we try to measure DNMTs enzyme expression level to demonstrate reduction of the DNA methylation are results of the reduction of DNMTs expression. Moreover, the results from this study can contribute to research assessing whether the beneficial and potential of DNMTs expression level as biomarker or therapeutic target for patients with JIA.
Material and methodsAll patients, which individuals in this study, were recruited from the division of pediatric rheumatology at Mofid children's Hospital in Tehran (Iran). JIA Patients were from all ethnicities, under the age of 16 and diagnosed according to the ACR criteria by a rheumatologist. The control individuals were chosen from healthy children who came to the hospital for checkup or minor surgical procedure without any autoimmune disease. The mean age of young patients and controls was 7.5 years old.
Peripheral blood mononuclear cells (PBMCs) were isolated from 3 ml fresh blood as soon as possible using Lymphoperp (Inno-Train, cat no 002041600, Germany). RNA was extracted using total RNA extraction kit (Pars Tous Biotech, cat no A101231, IR.IRAN). Five micrograms of total RNA samples were reverse transcribed to cDNA using the cDNA synthesis kit (iNtRON Biotechnology, cat no 25081, Korea). Then quantitative real time PCR was used to quantify expression levels of DNMT1 (NM_001130823.3), DNMT3a (NM_175629.2) and DNMT3b (NM_006892.3) genes. Primers used for the analysis of DNMTs expression are shown in Table 1. Primers were designed to analyze the expression of all DNMTs isoforms and situated on separate exons to avoid genomic DNA amplification.
Expression was measured using a quantitative real-time PCR machine (on Rotor gene 6000 Corbett Real Time PCR System) with SYBR Green PCR Master Mix (Ampliqon, cat no A324406, Denmark). The annealing temperature was 56 °C and the real-time PCR involved 35 cycles of amplification. ACTB gene (NM_007393) was used as an endogenous control to normalize expression data. Relative gene expression and fold changes were calculated using the following formulae14:
Statistical methodsData were normalized by using the log transformation. t-test was used to compare the relative expression of DNMTs between young patients and healthy controls. P < 0.05 for t was considered statistically significant. SPSS 21 and REST 2009 were used for this analysis.
Ethical approvalThe study was approved by the ethics committee of Shahid Beheshti University and Mofid children's Hospital. Informed written consent was obtained from all patients and/or their parents prior to their enrollment in this study.
28 JIA patients and 28 age-(9.3 ± 4.5, 9.1 ± 5.3 respectively) and sex-(9/19, 12/16 respectively) matched healthy control were enrolled in the study (more information about patients and healthy control are available in Table 2) and to analyze the expression of DNA methyltransferases in PBMCs of patients with JIA and healthy controls, PBMCs from individual patients and individual healthy controls were isolated. Total RNA was extracted and cDNA synthesis was carried out. RNA transcripts were quantified using quantitative real time PCR. The relative mRNA expression of DNMTs in JIA patients were compared with that in control individuals. Our results show that the expression of DNMT1 and DNMT3a were significantly reduced in JIA patients compared to healthy individuals (P = 0.014 and P = 0.024, respectively) but DNMT3b expression reduction was not significant (P = 0.104). DNMT1 expression was reduced by 7 folds and the expression of DNMT3a was reduced by 5.5 folds in JIA patients (Fig. 1).
Demographic and clinical characteristics of enrolled individuals.
| Parameters | JIA patients n = 28 | Control individuals n = 28 |
|---|---|---|
| Sex (male/female) | 9/19 | 12/16 |
| Age (years) | 9.3 ± 4.5 | 9.1 ± 5.3 |
| age of disease onset | 8 ± 5.3 | – |
| Female age (years) | 10.7 ± 4.4 | 11 ± 4.7 |
| Male age (years) | 6.2 ± 3.5 | 6.2 ± 4.6 |
| JIA subtype (oligo/poly) | 21/7 | – |
| Female JIA subtype (oligo/poly) | 14/5 | – |
| Male JIA subtype (oligo/poly) | 7/2 | – |
| ESRa | 32.9 ± 17 | – |
| Corticosteroids treatment (n) | 18 | – |
| Methotrexate treatment (n) | 28 | – |
| Vitamin D intake (n) | 25 | – |
Data are means ± SD.
Our results show that the expression of DNMT1 was reduced by 12.5 folds in affected males compared with control males and by 5.2 folds in affected females compared with control females. The expression of DNMT3a was reduced by 14.2 folds in males and by 3.5 folds in females (Fig. 2). The expression of DNMT3b was reduced by 6 folds in affected males and by 1.5 folds in affected females compared with healthy individuals. These results indicate that the expression of DNA methyltransferases are more dramatically decreased in young male patients compared with healthy male controls. Further studies would be required to confirm these results and help understand the cause and consequence of this observation.
DNA methyltransferases are more severely reduced in young affected males. JIA patients and healthy controls are separated based on their sex and the expression of DNA methyltransferases in young affected males and females were compared with healthy individuals. Relative gene expression data are shown.
It has been shown that DNMT1, DNMT3a, and DNMT3b have different functions, even though they all have highly conserved motives for methyltransferase activity.15 Changing gene expression is known to be one of the major functions of DNA methylation. Different methylation in several DNA loci in patients affected with JIA and change in methylations in IL32 and MRPL28 genes were reported.16 Previous studies have shown that IL32 expression is increased in patients with JIA and it is associated with a decrease in DNA methylation at this gene compared to controls.13 Reduced methylation, particularly in CpG rich gene promoters, is associated with elevated gene expression.17 Genetic disruption of DNMT1 and DNMT3b eliminates methyltransferase activity of both enzymes and thereby reduces DNA methylation up to 95 %.18 During maturation of immune cells DNMT1,3a and 3b are upregulated19 to regulate the expression of several genes such as ILs, TNF20 and IFNs.21,22 In this study, we analyzed the expression of DNMTs in the PBMCs of patients with JIA. Our results show that the expression of DNMTs in circulating PBMC in patients with JIA is reduced. DNMT1 and DNMT3a are significantly reduced in JIA patients whereas DNMT3b expression reduction is not significant. Given that DNMT1, DNMT3a and 3b are the only methyltransferases in mammals, these results suggest that the genome of immune cells in JIA patients is hypomethylated compared to the genome of immune cells in healthy individuals. A previous study found that various genes related to innate immunity, including several members of the ILs are overexpressed in autoimmune disease.23 The upregulation of ILs may be caused by promoter hypo methylation due to the downregulation of DNMTs. Reduction of DNMTs can affect the expression of several genes involved in inflammation and cell proliferation24 causing auto reactivity of T cells.
JIA is more common in young females than in young males; but males are more severely affected by the disease compared to females.25 We next asked, whether the expression of DNMTs in JIA patients was differently altered in male and female patients. To answer this question, JIA patients were separated based on their sex (Table 1) and the relative expression of these genes in young female and male patients were compared with that in young female and male controls, respectively. We found that in young affected females only DNMT1 show a significant reduction while in young affected males all three enzymes are significantly reduced. There are few potential explanations for the observed difference between affected males and females in terms of the expression of de novo methyltransferases. Female hormones affect DNMTs expression. In fact, it has been shown that progesterone and estrogen downregulate de novo methyltransferases.26 Furthermore, it has been shown that methotrexate (MTX), which is used to treat JIA patients, decreases female hormonal levels.27 Given that our patients were treated with MTX and that several female individuals included in this study were at puberty age, one can hypothesize that methotrexate treatment in young female patients ‘causes’ hormone reduction which in turn leads to an increase in de novo methyltransferases expression. On the other hand, in healthy females at puberty age, the increase in female hormones leads to a decrease in de novo methyltransferases expression. Hence, we failed to find a significant difference between the expression of de novo methyltransferases in JIA female patients and healthy female controls.
The most important limitation of this study is low samples in each subtypes of JIA. Because of this limitation, sample number of the each subtype are not enough for comparing of the expression of DNMTs between them. Based on this fact and previous study which show polyarticular and oligoarticular are been result of same immune responds,28 we considered them as same. In some cases, the parents did not agree to donor the blood of their children. In addition, we cannot take more than 3 ml blood from children, and always parents just agree to donor blood one time from their children. Therefore, blood is miss or anything happen during experiment, the case is not available again.
In conclusion, this study shows that DNMTs are reduced in JIA patient and the expression of DNMTs is severely affected in young male JIA patients. Further investigation is required to explain the difference between JIA affected females and males in terms of DNMTs expression. A prospective cohort study, the study of untreated young patients and the separate examination of different types of blood cells is required. It would also be interesting to analyze DNA methylation status of inflammatory genes in PBMC of JIA patients.
Conflicts of interestThe authors have no conflict of interest to declare.
The authors would like to thank Pediatric Infections Research Center of Shahid Beheshti University of Medical Sciences’ staff member for their support and assistance with this project. Special thanks also go to the Mofid children hospital pediatric rheumatology department’s staff and the blood donor, who voluntarily donate their blood to help save lives.