To characterize OXA-48 carbapenemase-producing Klebsiella pneumoniae strains isolated after an increase in carbapenem resistance in Catalonia.
MethodologyK. pneumoniae identification, antimicrobial susceptibility studies, the Modified Hodge Test method, amplification of antimicrobial resistance genes (against β-lactamases, quinolones and aminoglycosides), molecular typing (by PFGE and MLST), conjugation assays, plasmid characterization (PBRT-PCR and Southern blot), a description of mobile genetic elements and statistical analysis were done.
ResultsOXA-48 was the only carbapenemase detected, with a prevalence of 1.9%. The blaOXA-48 gene was located in an IncL conjugative plasmid of 62kb and integrated into the transposons Tn1999.2 (91.7%) or Tn1999.1. Five PFGE profiles (A to E) were found, which exactly matched the MLST: ST101, ST17, ST1233, ST14 and ST405, respectively. ST1233 is described here for the first time. K. pneumoniae OXA-48-producing strains were also CTX-M-15 carriers, some producing OXA-1 and TEM-1 penicillinases. The acquired qnrB66 and qnrB1 and aac(3′)-IIa, aac(6′)-Ib genes were also identified.
ConclusionThe K. pneumoniae ST405 clone has played an important role in the growing prevalence of OXA-48 in Catalonia. All clones described preserved the blaOXA-48 genetic environment and mobile genetic elements (Tn1999). Notably, the three strains with minor sequence types in this study are not multiresistant strains. These strains are expanding in elderly patients (average age of 76 years) with serious underlying diseases, mainly women (61.2%).
El objetivo de este estudio fue caracterizar las cepas de Klebsiella pneumoniae productoras de carbapenemasa OXA-48 aisladas tras observar un aumento de estos aislados resistentes a los carbapenémicos en Cataluña.
MétodosSe realizó la identificación de K. pneumoniae, estudios de sensibilidad antimicrobiana, el test de Hodge modificado, amplificación de genes de resistencia antimicrobiana (contra β-lactamasas, quinolonas y aminoglucósidos), tipificación molecular (por PFGE y MLST), ensayos de conjugación, caracterización de plásmidos (PBRT-PCR y Southern blot), descripción de los elementos genéticos móviles y el análisis estadístico.
ResultadosOXA-48 fue la única carbapenemasa presente, con una prevalencia del 1,9%. El gen blaOXA-48 se localizó en un plásmido conjugativo IncL de 62kb e integrado en los transposones Tn1999.2 (91,7%) o Tn1999.1. Se encontraron 5 perfiles diferentes de PFGE (A a E), que tenían una concordancia exacta con el MLST: ST101, ST17, ST1233, ST14 y ST405, respectivamente. El ST1233 se describe aquí por primera vez. Las cepas productoras de K. pneumoniae OXA-48 también fueron portadoras de CTX-M-15 y algunas de ellas productoras también de penicilinasas OXA-1 y TEM-1. Los genes adquiridos qnrB66 y qnrB1 y aac(3’)-IIa, aac(6’)-Ib también se identificaron.
ConclusiónEl clon K. pneumoniae ST405 tiene un papel importante en la creciente prevalencia de OXA-48 en Cataluña. Todos los clones descritos preservaron el entorno genético de blaOXA-48, así como los elementos genéticos móviles (Tn1999). Notablemente, las 3 cepas con tipos de secuencia menos prevalentes en este estudio no son cepas multirresistentes. Además, la expansión de estas cepas con blaOXA-48 se está produciendo en pacientes de edad avanzada (promedio de edad de 76 años), la mayoría mujeres (61,2%) con enfermedades subyacentes graves.
The emergence and spread of carbapenem-resistant Enterobacteriaceae due to carbapenemase production is a serious public health problem worldwide. Colistin or tigecycline are last-resort antibiotics against these carbapenemase-producing Enterobacteriaceae,1 but some resistant strains have been described.
Class A, B and D carbapenemases have been reported in Enterobacteriaceae worldwide since 1993,2,3 but were not described in Spain until 2003.4 VIM and IMP (class B) were initially the most frequent, but they have since then declined, and OXA-48 (class D), first documented in Spain in 2009, is currently the most prevalent.5
The OXA-48 carbapenemase exhibits strong penicillin-hydrolysing activity and weak activity against carbapenems. Derivatives such as OXA-163 (first described in Klebsiella pneumoniae), OXA-247 (K. pneumoniae) and OXA-405 (Serratia marcescens) hydrolyse penicillins, ceftazidime and cefotaxime, but as their carbapenem-hydrolysing activity is far lower than OXA-48 they are barely considered as carbapenemases.1–3
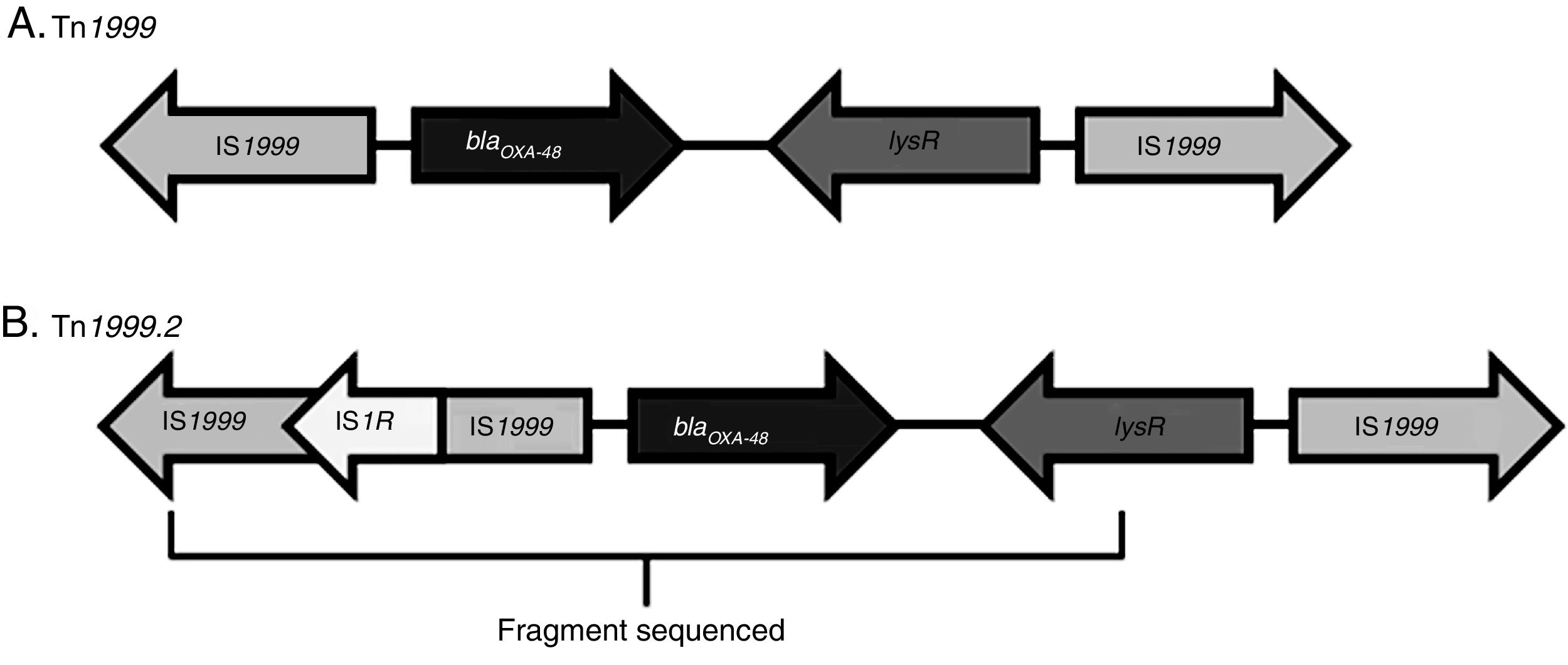
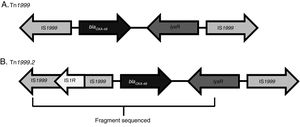
An OXA-48-producing K. pneumoniae strain was first identified in 2001 in Istanbul, with reservoirs becoming established in North Africa and the Mediterranean area. It has now spread through the rest of Europe, Asia and America.6,7 The dissemination of this enzyme could be explained by two factors. First, blaOXA-48 has been described in an IncL plasmid in Enterobacteriaceae as well as in both non-fermenters (Acinetobacter baumannii and Pseudomonas aeruginosa).1 Second, blaOXA-48 gene is part of the transposon Tn1999, which is inserted in the tir gene of the plasmid IncL. Different Tn1999 variants are known (Tn1999.2, Tn1999.3 and Tn1999.4),8 varying in the presence or not of the insertion sequence IS1R1 or the transposon Tn2015, which also contains blaCTX-M-15. On the other hand, the insertion of Tn1999 in the tir gene of this plasmid has been associated with an enhanced conjugation performance and could contribute to the high diffusion of this plasmid type and its resistance genes.
Enterobacteriaceae strains carrying the OXA-48-carbapenemase can also co-express extended-spectrum β-lactamases (ESBLs) such as CTX-M-15 and SHV-12,9 or acquired AmpC β-lactamases (acAmpC) such as DHA-1,4 mainly in different plasmids; strains resistant to quinolones, cotrimoxazole and aminoglycosides have also been described.2,10
In a national study conducted in 2009,4 we observed a very low prevalence of carbapenemase-producing Enterobacteriaceae in Catalonia (0.04%). However, in 2012, hospitals in the Barcelona area reported an increase in K. pneumoniae strains resistant to third generation cephalosporins and ertapenem and suspected of expressing carbapenemases. The aim of this study was to explain this growing prevalence by characterizing the β-lactamases involved in this resistance phenotype and establishing the genetic relationships between strains.
Material and methodsEthicsThe study was approved by the Ethical Review Committee of the Institut de Recerca de l’Hospital de la Santa Creu i Sant Pau.
Strains and patientsIn a prospective study in 2012 (January to December) involving 12 hospitals in Catalonia, K. pneumoniae isolates resistant to β-lactams were collected, excluding those with a natural resistance pattern. Epidemiological data on patient gender, age, chronic diseases and treatment were collected in parallel. Only one strain per patient was included. The selected strains were resistant to any of the following β-lactams: cephalotin, cefoxitin, cefuroxime, cefotaxime, ceftazidime, cefepime, ertapenem, imipenem, aztreonam, amoxicillin/clavulanic acid and/or piperacillin/tazobactam.
Each hospital carried out identification and antibiotic susceptibility tests and provided epidemiological patient data. The participating hospitals were: Hospital Municipal de Badalona (HMB), Hospital de Barcelona (HB), Corporació de Salut del Maresme I la Selva (HC), Hospital General de Granollers (HGG), Hospital General Universitari de Catalunya at Sant Cugat del Vallès (HGC), Hospital General de L’Hospitalet (HGH), Hospital Sant Joan de Déu at Manresa (HSJDD), Hospital Sant Joan Martorell (HMLL), Hospital de Mataró (HM), Hospital Universitari de Vic (HGV), Hospital Universitari de Sant Joan de Reus (HUSJR) and Hospital de la Santa Creu i Sant Pau (HSCSP) (covering a population of approximately 250,000 inhabitants).
The patients were classified in three categories: nosocomial (the infection occurring after 48h of hospital admission), healthcare centre (resident in a healthcare centre) and community (no recent contact with a medical environment).
Bacterial identification and antimicrobial susceptibility testingK. pneumoniae identification and antimicrobial susceptibility tests were performed in each hospital following routine laboratory methods, either manual or automated [MicroScan WalkAway (Siemens) and Vitek system (bioMérieux, Marcy l’Etoile, France)]. The susceptibility pattern to β-lactams, quinolones and aminoglycosides was obtained by the disc-diffusion method (Rosco Diagnostica A/S, Taastrup, Denmark) following Clinical and Laboratory Standards Institute (CLSI) criteria, as used routinely in the laboratory.11 The antibiotics used were: ampicillin (AMP), piperacillin (PIP), amoxicillin/clavulanic acid (AMC), piperacillin/tazobactam (TZP), cephalotin (CEF), cefoxitin (FOX), cefuroxime (CXM), cefotaxime (CTX), ceftazidime (CAZ), aztreonam (ATM), cefepime (FEP), ertapenem (ERT), imipenem (IMP), kanamycin (K), gentamicin (G), tobramycin (T), amikacin (A), netilmicin (Nt), neomycin (Nm), nalidixic acid (NAL), ciprofloxacin (CIP), and cotrimoxazole (SXT).
The then recommended Modified Hodge Test (MHT) was performed to detect carbapenemase activity, using imipenem according to CLSI criteria.11 The strains selected for analysis were resistant to any of the studied β-lactams. Strains with positive or weakly positive MHT results were included for the molecular characterization of carbapenemase resistance mechanisms.
Amplification of antimicrobial resistance genesThe polymerase chain reaction (PCR) was used to detect the following genes in all studied strains according to their resistance phenotype: carbapenemases (blaOXA-48, blaVIM, blaSPM, blaIMP, blaGIM, blaSME, blaNMC, blaKPC, blaIMI and blaGES, blaNDM), ESBLs (blaTEM, blaSHV, blaCTX-M), acquired AmpC genes (acAmpC) (blaACC, blaCIT, blaEBC, blaDHA, blaFOX, blaMOX) and the penicillinase blaOXA-1, quinolones (qepA and qnrA, qnrB, qnrS, qnrC, qnrD) and aminoglycoside modifying enzymes (AME) (aac(3′)-IIa, aac(6)-Ib, aph(3′)-Ia, ant(2′)-Ia and aac(2′)-Ia).4,12,13
The blaOXA-48 genetic platform was determined by PCR using specific primers for IS1999, lysR and IS1R of Tn1999 and Tn1999.26,14 in all blaOXA-48-carrying strains.
All amplicon sequences were obtained by Sanger sequencing in an external genome service enterprise (Macrogen Europe, Amsterdam, The Netherlands).
Molecular typing of blaOXA-48-carrying K. pneumoniae strainsPulsed-field gel electrophoresis (PFGE) with genomic DNA digestion by the enzyme XbaI was performed with the CHEF-DRIII system (Bio-Rad, Hemel Hempstead, UK) and analyzed by BioNumerics 6.6 (Applied-Maths, Ghent, Belgium).2,12,15 The relatedness was calculated by the unweighted-pair group method using an average linkage (UPGMA) algorithm, with band similarity calculated using the Dice coefficient with a 2% optimization value and 1% tolerance. PFGE patterns were interpreted as previously described.11,12 MultiLocus Sequence Typing (MLST) was performed according to the Pasteur Institute website (www.pasteur.fr/recherche/genopole/P8/mlst)15 in 11 strains selected according to their PFGE pattern (A-E). For cluster E, we selected the majority and minority patterns consulting the resistance pattern.
Conjugation assayConjugation was performed in the aforementioned 11 isolates. Rifampicin-resistant-GFP E. coli VA6190 was used as a recipient,11 which also expresses a green fluorescent protein (GFP) marker. Luria-Bertani broth (LB) was used for the conjugation or, if LB results were negative, solid mating with HA 45μm-pore-size filters (Millipore, Billerica, MA). Putative transconjugants were selected on LB agar supplemented with ceftazidime (10mg/L) and rifampicin (100mg/L), and exposed to UV illumination to check for GFP fluorescence. The transfer frequency was expressed as the ratio of transconjugants to total recipient cells. The presence of blaOXA-48 in the transconjugants was confirmed by PCR.
Plasmid typingPlasmid Identification was performed in 37 strains by a homemade PCR-based replicon typing (PBRT-PCR) method based on Carattoli et al. (2009).16 The studied strains represented each PFGE subcluster, adding some strains with the same PFGE pattern but different resistance patterns. Poirel et al. (2012)17 observed that PBRT-PCR did not detect a blaOXA-48-carrying IncL/M replicon. Carattoli et al. (2015)18 described specific primers to detect the blaOXA-48-carrying IncL plasmid, which distinguished IncL from IncM. We used these primers to describe the plasmid in our OXA-48-producing K. pneumoniae strains.
The plasmid localization of the bla genes was determined in the same 11 strains by PFGE using S1 nuclease and Southern blot hybridization methods, as previously described.12 The probes were obtained by the DIG Probe Synthesis Kit (Sigma–Aldrich) using primers corresponding to the bla and replicon genes.
Statistical analysisCategorical variables were compared by the χ2 test and continuous variables by the Student's t-test or Mann–Whitney test. P-values of <0.005 were considered statistically significant. The software GraphPad Prism (GraphPad Software, Inc., CA, USA) was used for the analyses.
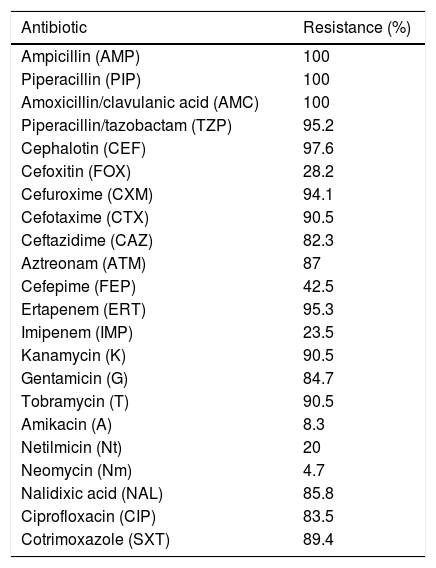
ResultsBacterial isolates and susceptibility dataBetween January and December 2012, we collected a total of 3901 K. pneumoniae isolates resistant to β-lactams, not including those with a natural resistance pattern. Among these, 171 (4.38%) gave positive MHT results, including strains with only a weak growth around the streak. And among these171 strains, 98 (57.3%) were resistant to ertapenem and 22 (12.8%) to imipenem. After the PCR and sequencing, 85 strains gave positive results for blaOXA-48, nine of them from faecal carriers. These 85 strains were from eight out of the 12 participating hospitals. Thus, the prevalence of OXA-48-producing K. pneumoniae strains in Catalonia was 1.9% (subtracting the nine faecal carriers). No other targeted carbapenemases were detected. The 85 OXA-48-producing strains were resistant to ampicillin, piperacillin and the association amoxicillin/clavulanic acid. Only 23.5% were resistant to imipenem, whereas 95.3% were resistant to ertapenem, which is the most sensitive drug for OXA-48-producing strain detection. Most strains were resistant to cephalosporins, in some cases remaining susceptible to cefoxitin and cefepime (Table 1). These different phenotypes were due to the presence of an ESBL, as 89.4% co-expressed the CTX-M-15, together with the penicillinases OXA-1 (94.1%) and TEM-1 (85.4%). All 85 isolates also expressed the chromosomal penicillinase SHV characteristic of this species: in 77.64% we detected blaSHV-76, 20% blaSHV-1, 1.1% blaSHV-11 and 1.1% blaSHV-42.
Antibiotic resistance in OXA-48-producing K. pneumoniae strains.
| Antibiotic | Resistance (%) |
|---|---|
| Ampicillin (AMP) | 100 |
| Piperacillin (PIP) | 100 |
| Amoxicillin/clavulanic acid (AMC) | 100 |
| Piperacillin/tazobactam (TZP) | 95.2 |
| Cephalotin (CEF) | 97.6 |
| Cefoxitin (FOX) | 28.2 |
| Cefuroxime (CXM) | 94.1 |
| Cefotaxime (CTX) | 90.5 |
| Ceftazidime (CAZ) | 82.3 |
| Aztreonam (ATM) | 87 |
| Cefepime (FEP) | 42.5 |
| Ertapenem (ERT) | 95.3 |
| Imipenem (IMP) | 23.5 |
| Kanamycin (K) | 90.5 |
| Gentamicin (G) | 84.7 |
| Tobramycin (T) | 90.5 |
| Amikacin (A) | 8.3 |
| Netilmicin (Nt) | 20 |
| Neomycin (Nm) | 4.7 |
| Nalidixic acid (NAL) | 85.8 |
| Ciprofloxacin (CIP) | 83.5 |
| Cotrimoxazole (SXT) | 89.4 |
These strains were also resistant to aminoglycosides (90.5%), cotrimoxazole (89.4%) and ciprofloxacin (83.5%). Only four strains remained susceptible to all aminoglycosides and quinolones tested. Regarding aminoglycoside resistance, we found nine different phenotype patterns: KTG (64.7%), KTGN (14.1%), KTGNm (4.7%), KTA (1.1%), G (2.3%), KTAGN (1.1%), KTAN (3.5%), KT (2.3%) and KTN (1.1%). The AMEs present in these strains were AAC(3′)-IIa (83.5%), AAC(6′)-Ib (81.2%), APH(3′)-Ia (3.5%), AAC(2′)-Ia (2.4%) and ANT (2′)-Ia (1.2%). Finally, 88.2% carried QnrB and in one case QnrS (Table 2).
Antimicrobial resistance genes present in the different MLST and PFGE profiles of the 85 OXA-48-producing K. pneumoniae strains.
| ST | PFGE | n | β-Lactamase genes | AME | Qnr |
|---|---|---|---|---|---|
| ST101 | A1 | 6 | blaCTX-M-15, blaOXA-1, blaTEM-1, blaSHV-1 (100%a) | aac(3′)-IIa, aac(6′)-Ib (66.6%) aac(3′)-IIa, aac(6′)-Ib, aph(3′)-Ia (33.3%) | qnrB66 qnrS (50%) |
| A2 | 9 | blaCTX-M-15, blaOXA-1, blaSHV-1 (55.5%) blaCTX-M-15, blaOXA-1, blaTEM-1, blaSHV-1 (44.4%) | aac(3′)-IIa, aac(6′)-Ib (66.6%) aac(3′)-IIa, aac(6′)-Ib, aac(2′)-Ia (11.1%) aac(3′)-IIa, aac(6′)-Ib, ant(2′)-Ia, aac(2′)-Ia (11.1%) aac(3′)-IIa, aac(6′)-Ib, aph(3′)-Ia (11.1%) | qnrB66 (45.5%) | |
| A3 | 1 | blaCTX-M-15, blaOXA-1, blaTEM-1, blaSHV-1 | aac(3′)-IIa, aac(6′)-Ib, aph(3′)-Ia | qnrB1 | |
| ST17 | B | 1 | blaSHV-11 | – | – |
| ST1233 | C | 1 | blaSHV-42 | – | – |
| ST14 | D | 1 | blaSHV-1 | – | – |
| ST405 | E1 | 11 | blaOXA-1, blaTEM-1, blaSHV-76 (9%) blaOXA-1, blaSHV-76 (9%) blaCTX-M-15, blaOXA-1, blaSHV-76 (9%) blaCTX-M-15, blaTEM-1, blaSHV-76 (18%) blaCTX-M-15, blaOXA-1, blaTEM-1, blaSHV-76 (55%) | aac(3′)-IIa, aac(6′)-Ib (36.3%) aac(3′)-IIa (36.3%) aac(6′)-Ib (18.1%) | qnrB66 (91%) |
| E2 | 1 | blaCTX-M-15, blaOXA-1, blaTEM-1, blaSHV-76 | aac(3′)-IIa, aac(6′)-Ib | qnrB66 | |
| E3 | 1 | blaOXA-1, blaSHV-76 | aac(6′)-Ib | qnrB66 | |
| E4 | 3 | blaCTX-M-15, blaOXA-1, blaTEM-1, blaSHV-76 | aac(3′)-IIa, aac(6′)-Ib | qnrB66 | |
| E5 | 30 | blaCTX-M-15, blaOXA-1, blaTEM-1, blaSHV-76 | aac(3′)-IIa, aac(6′)-Ib (90%) aac(3′)-IIa (10%) | qnrB66 (96%) | |
| E6 | 2 | blaCTX-M-15, blaOXA-1, blaTEM-1, blaSHV-76 (50%) blaOXA-1, blaSHV-76 (50%) | aac(3′)-IIa, aac(6′)-Ib (50%) aac(6′)-Ib (50%) | qnrB66 | |
| E7 | 1 | blaCTX-M-15, blaOXA-1, blaTEM-1, blaSHV-76 | aac(3′)-IIa, aac(6′)-Ib | qnrB66 | |
| E8 | 1 | blaCTX-M-15, blaOXA-1, blaTEM-1, blaSHV-76 | aac(3′)-IIa, aac(6′)-Ib | qnrB66 | |
| E9 | 1 | blaCTX-M-15, blaOXA-1, blaTEM-1, blaSHV-76 | aac(3′)-IIa, aac(6′)-Ib | qnrB66 | |
| E10 | 1 | blaOXA-1, blaTEM-1, blaSHV-76 | aac(6′)-Ib | qnrB66 | |
| E11 | 1 | blaCTX-M-15, blaOXA-1, blaTEM-1,blaSHV-76 | aac(3′)-IIa, aac(6′)-Ib | qnrB66 | |
| E12 | 5 | blaCTX-M-15, blaOXA-1, blaTEM-1,blaSHV-76 (100%) | aac(3′)-IIa, aac(6′)-Ib | qnrB66 | |
| E13 | 1 | blaCTX-M-15, blaOXA-1, blaTEM-1,blaSHV-76 | aac(3′)-IIa, aac(6′)-Ib | qnrB66 | |
| E14 | 1 | blaCTX-M-15, blaOXA-1, blaTEM-1, blaSHV-76 | aac(3′)-IIa, aac(6′)-Ib | qnrB66 | |
| E15 | 1 | blaCTX-M-15, blaOXA-1, blaTEM-1, blaSHV-76 | aac(3′)-IIa, aac(6′)-Ib | qnrB66 | |
| E16 | 1 | blaCTX-M-15, blaOXA-1, blaTEM-1, blaSHV-76 | aac(3′)-IIa, aac(6′)-Ib | qnrB66 | |
| E17 | 1 | blaOXA-1, blaTEM-1, blaSHV-76 | aac(3′)-IIa, aac(6′)-Ib | qnrB66 | |
| E18 | 3 | blaCTX-M-15, blaOXA-1, blaTEM-1, blaSHV-76 | aac(3′)-IIa, aac(6′)-Ib | qnrB66 |
As mentioned above, 86 out of 171 positive MHT strains (50.3%) were non-carbapenemase-producing. Among these isolates, 17 (19.7%) were resistant to ertapenem and 1 (1%) to imipenem. To explain the positive MHT results, we checked for the presence of ESBLs and acAmpC. The PCR results revealed that 47 strains (54.6%) carried an acAmpC (80.8% DHA, 10.6% ACC and 8.6% CMY), 21 (24.4%) carried an ESBL (CTX-M-1-type) and 14 (16.3%) co-expressed both ESBL and acAmpC (64.3% CTX-M-1-type+DHA, 14.3% CTX-M-9-type+DHA, 14.3% CTX-M-1-type+ACC and 7.1% CTX-M-1-type with CMY). Finally, four strains (4.6%) did not show any studied acAmpC or ESBL. The 86 strains without carbapenemase production were excluded from further studies.
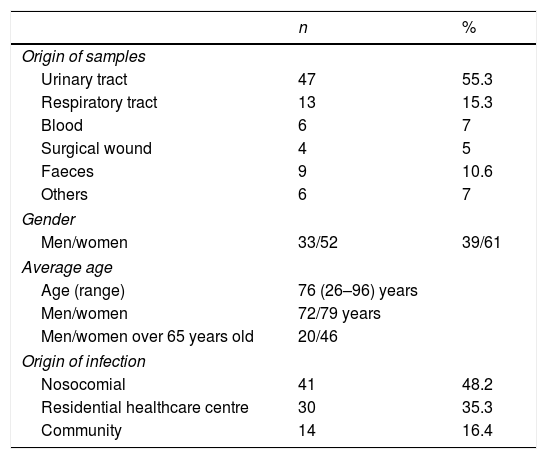
Clinical and molecular epidemiological dataThe origin of the 85 OXA-48-producing strains was: 47 (55.3%) from urine samples, 13 (15.3%) from respiratory tract samples, six (7%) from blood, four (4.7%) from surgical wounds, six (7%) from other samples (vaginal discharge, peritoneal fluid, sore, cellulitis, bile, post-operative ulcer) and nine (10.6%) were isolated from faeces (Table 3).
Clinical data of the 85 patients with OXA-48-producing K. pneumoniae.
| n | % | |
|---|---|---|
| Origin of samples | ||
| Urinary tract | 47 | 55.3 |
| Respiratory tract | 13 | 15.3 |
| Blood | 6 | 7 |
| Surgical wound | 4 | 5 |
| Faeces | 9 | 10.6 |
| Others | 6 | 7 |
| Gender | ||
| Men/women | 33/52 | 39/61 |
| Average age | ||
| Age (range) | 76 (26–96) years | |
| Men/women | 72/79 years | |
| Men/women over 65 years old | 20/46 | |
| Origin of infection | ||
| Nosocomial | 41 | 48.2 |
| Residential healthcare centre | 30 | 35.3 |
| Community | 14 | 16.4 |
The demographic data collected from the 85 patients with OXA-48-producing K. pneumoniae infection showed that 52 were female (61.2%) of an average age of 76 years (range 26–98 years). There was no significant difference between genders (P>0.005). In 41 (48.2%) cases the infection had a nosocomial origin, 30 (35.3%) were related to a healthcare centre, and the remaining 14 (16.5%) were acquired in the community (Table 3).
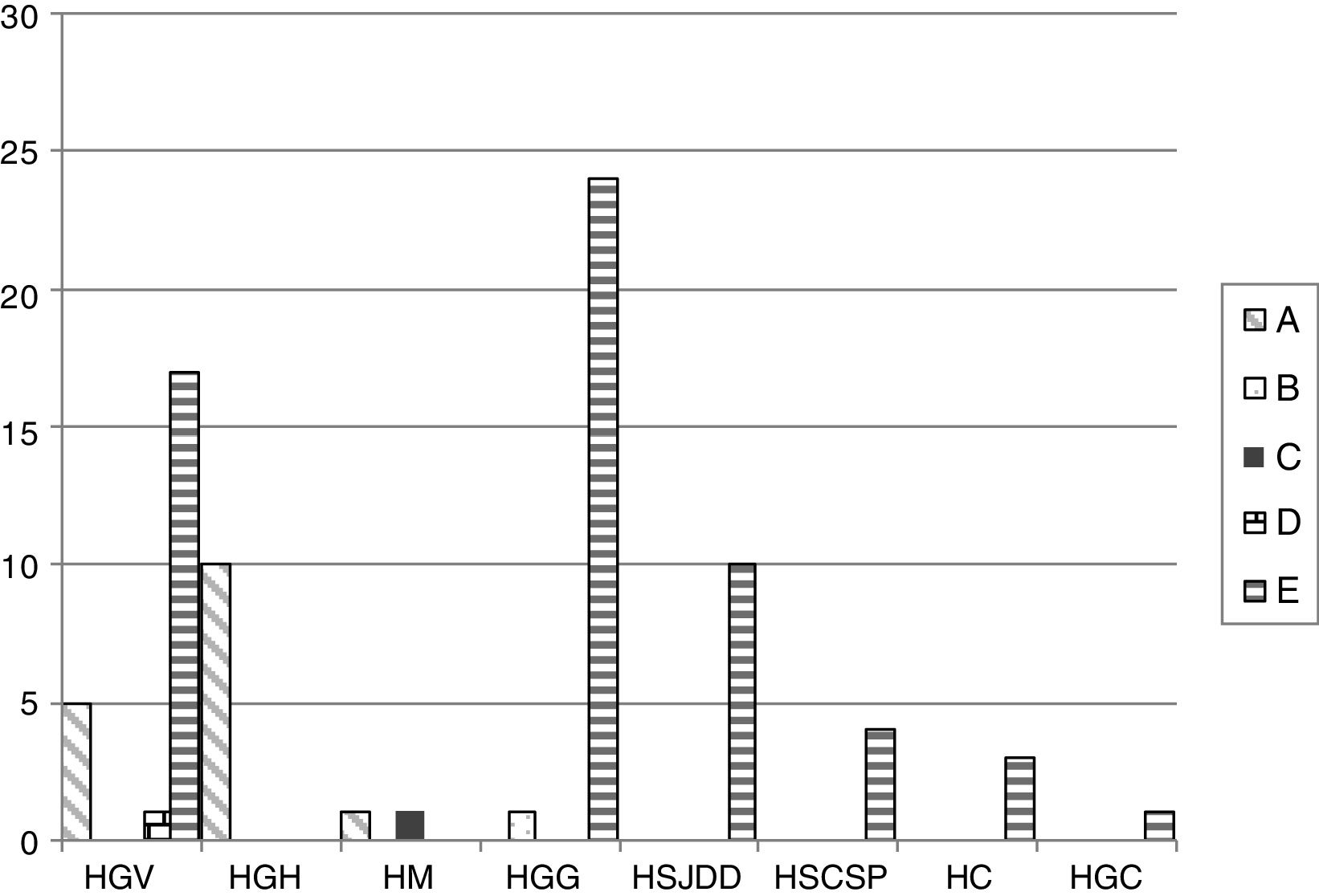
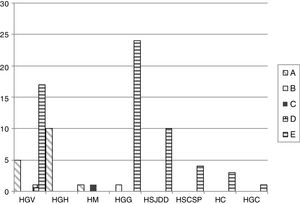
The PFGE analysis revealed five well-defined clusters (A–E), which were subdivided in multiple subclusters, with 76% homology between the main clusters. Cluster A, B, C, D and E represented 18.8%, 1.1%, 1.1%, 1.1% and 77.6% of all isolates, respectively. Clusters A and E showed subclusters A1-3 and E1-18. The cluster distribution is depicted in Fig. 1.
Distribution of the different PFGE clusters by hospital. HC (Consorci de Salut del Maresme), HGC (Hospital General Universitari de Catalunya), HGG (Hospital General de Granollers), HGH (Hospital General de l’Hospitalet), HGV (Consorci hospitalari de Vic), HM (Hospital de Mataró), HSCSP (Hospital de la Santa Creu i Sant Pau) and HSJDD (Hospital Sant Joan de Déu de Manresa).
By MLST, the five PFGE clusters corresponded to five different K. pneumoniae sequence types (STs) as follows: cluster A belonged to ST101 (n=16); B to ST17 (n=1); C to ST1233 (n=1); D to ST14 (n=1) and E to ST405 (n=66). ST1233 is described here for the first time and is a single-locus variant (SLV) of ST540 in the gapA gene. Cluster E, which corresponds with ST405, was isolated in six of the eight OXA-48-detecting hospitals; cluster A, all ST101, was present in three of the eight hospitals. The minor clusters B (ST17), C (ST1233) and D (ST14), with one strain each, were from HGG, HM and HGV, and were isolated in different hospitals together with isolates belonging to major clones.
We observed that all OXA-48-producing K. pneumoniae strains, except those belonging to ST14, ST17 and ST1233, were resistant to multiple antimicrobial drugs (mainly aminoglycosides and quinolones) by acquiring resistance genes. blaOXA-48 seemed to be associated with blaOXA-1, aac(3′)-IIa, aac(6′)-Ib and qnrB, and in most cases with blaCTX-M-15 and blaTEM-1 (Table 2).
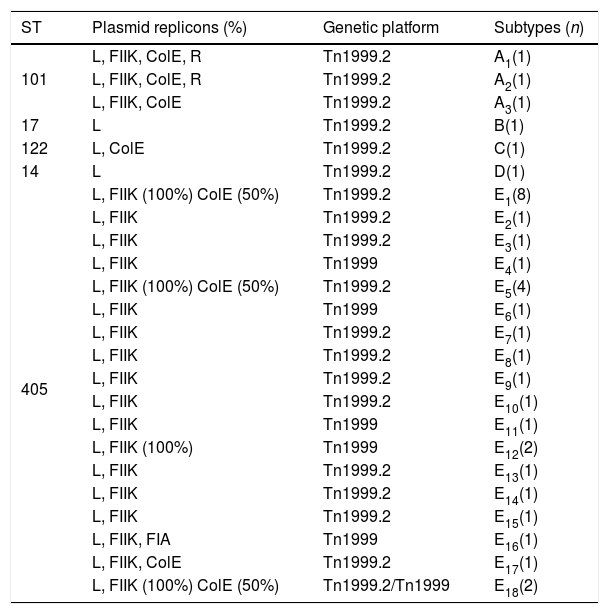
Plasmid characterization and the blaOXA-48-carrying genetic platformThe PBRT-PCR data show that the 37 selected strains carried a plasmid with an IncL plasmid, 96% out of the 37 a plasmid replicon FIIK and 33.3% a plasmid replicon ColE (Table 4). Two cluster A-ST101 strains also carried one plasmid replicon R and the strain of cluster E16-ST405 showed the plasmid replicon FIA. The strains belonging to ST17 and ST14 were the only ones without plasmid replicons other than IncL.
Plasmid typing (PBRT-PCR) in 37 OXA-48-producing K. pneumoniae strains.
| ST | Plasmid replicons (%) | Genetic platform | Subtypes (n) |
|---|---|---|---|
| 101 | L, FIIK, ColE, R | Tn1999.2 | A1(1) |
| L, FIIK, ColE, R | Tn1999.2 | A2(1) | |
| L, FIIK, ColE | Tn1999.2 | A3(1) | |
| 17 | L | Tn1999.2 | B(1) |
| 122 | L, ColE | Tn1999.2 | C(1) |
| 14 | L | Tn1999.2 | D(1) |
| 405 | L, FIIK (100%) ColE (50%) | Tn1999.2 | E1(8) |
| L, FIIK | Tn1999.2 | E2(1) | |
| L, FIIK | Tn1999.2 | E3(1) | |
| L, FIIK | Tn1999 | E4(1) | |
| L, FIIK (100%) ColE (50%) | Tn1999.2 | E5(4) | |
| L, FIIK | Tn1999 | E6(1) | |
| L, FIIK | Tn1999.2 | E7(1) | |
| L, FIIK | Tn1999.2 | E8(1) | |
| L, FIIK | Tn1999.2 | E9(1) | |
| L, FIIK | Tn1999.2 | E10(1) | |
| L, FIIK | Tn1999 | E11(1) | |
| L, FIIK (100%) | Tn1999 | E12(2) | |
| L, FIIK | Tn1999.2 | E13(1) | |
| L, FIIK | Tn1999.2 | E14(1) | |
| L, FIIK | Tn1999.2 | E15(1) | |
| L, FIIK, FIA | Tn1999 | E16(1) | |
| L, FIIK, ColE | Tn1999.2 | E17(1) | |
| L, FIIK (100%) ColE (50%) | Tn1999.2/Tn1999 | E18(2) |
PFGE and Southern blot studies revealed that that blaOXA-48 was present in all 85 OXA-48-producing strains in an approx. 62kb plasmid belonging to the IncL incompatibility group. These data were confirmed by conjugation assays. In all 11 conjugated strains we found OXA-48-producing E. coli transconjugants with a conjugation frequency between 1.3×10−5 and 5×10−7. All transconjugants with blaOXA-48 were resistant to penicillins, remaining susceptible to cefotaxime and ceftazidime and with a reduction in their susceptibility to ertapenem. In all cases the carbapenemase had transferred alone, as confirmed by PCR.
Southern blot experiments revealed that blaCTX-M-15 and blaOXA-1 were carried in FIIK plasmids of approx. 240Kb in ST101strains and approx. 290Kb in ST405.
The blaOXA-48-carrying genetic platform was related with the transposon Tn1999 (Table 4). Seventy-eight strains (91.8%) showed Tn1999.2 and seven the intact Tn1999. We sequenced the blaOXA-48 genetic surroundings in eleven randomly selected strains (Fig. 2) (GenBank accession numbers: KT265174, KT265175, KT265176, KT265177, KT265178, KT265179, KT265180, KT265181, KT265182, KT265183 and KT265173). In 10 cases a unique sequence between the IS1R element, upstream of blaOXA-48 and lysR, was found, with 100% homology with the Tn1999.2 sequence from pKpn-E1.Nr7 (KM406491.1). The remaining strain had 100% homology with K. pneumoniae E71T (KC335143.1) and pKPoxa-48N1 (KC757416.2). The only difference between the two groups was due to a transversion T-G at position 911, where the K. pneumoniae E71T (KC335143.1) and pKPoxa-48N1 (KC757416.2) sequences have a guanine.
DiscussionDetermining OXA-48-producing strain prevalence is hampered by the lack of a standard method.19 The MHT, in 2012 recommended by the CLSI for carbapenemase detection,1,11 is now considered unsuitable as a single screening method, being too unspecific and insensitive for metallo-β-lactamase detection. In this study, 51% of positive MHT strains did not express any carbapenemase. These false positives were due to the presence of ESBLs such as CTX-M-15, acAmpCs such as DHA or porin alterations, which may be involved in reduced susceptibility to carbapenems.12 A low affinity for carbapenems of some ESBLs has been described, specifically for the CTX-M-15 enzyme, which can hydrolyse ertapenem when highly expressed.20 The three OXA-48-producing K. pneumoniae strains without additional β-lactamases were only resistant to ampicillin, piperacillin and amoxicillin/clavulanic acid, remaining susceptible to ertapenem and imipenem. In contrast, all the OXA-48- and ESBL- or acAmpC-producing strains were resistant to penicillins, cephalosporins and ertapenem, and 23.5% also to imipenem.
Therefore, assuming that the phenotypic method might not be accurate to determine the prevalence of oxa-48 producing strains, prevalence was determined according to PCR results. The prevalence of OXA-48-producing-K. pneumoniae strains in 2012 in Catalonia was 1.9%, matching previous studies in some Spanish tertiary hospitals2,9 a 0.04% and 5.3% prevalence was described. A recent European survey of carbapenemase-producing Enterobacteriaceae (EuSCAPE),21 found a 69.8% prevalence of OXA-48-producing Klebsiella pneumoniae strains among 136 strains collected by Instituto de Salud Carlos III (Majadahonda, Spain).
The lack of standardization in detection methods affects not only prevalence data, but also the implementation of a policy to avoid the establishment of multiresistant strains,22 although new chromogenic plate methods have been described.
Like ESBL- or acAmpC-producing strains,4,12,13 OXA-48-producing K. pneumoniae isolates are also resistant to aminoglycosides, cotrimoxazole and ciprofloxacin. Nevertheless, the blaOXA-48-carrying IncL plasmids described to date (such as pkPoxa-48N1),18 responsible for OXA-48 dissemination, do not include any associated antimicrobial resistance genes. Our results show that blaCTX-M-15 is present in an IncFIIk plasmid, in agreement with other authors,23 together with quinolone and aminoglycoside resistance genes.24
The worldwide expansion of blaOXA-48 could be due to its association with Tn1999, located on a characteristic IncL plasmid.1 In turn, Tn1999 is inserted into the tir gene responsible for plasmid transfer inhibition. We found the increasing OXA-48 prevalence was also due to the expansion of the ST405 clone, present in all hospitals with OXA-48-producing K. pneumoniae except two. This major clone was originally identified in Casablanca (Morocco)25 in a K. pneumoniae strain carrying blaCTX-M-15, blaOXA-1 and blaTEM-1, but not blaOXA-48, and has been widely described in Europe, including Spain9 As described by Baquero et al.,26 this expansion could be due to colonization and transmission between particular hosts, which acquire antibiotic resistance and enhanced survival capacity. In this study, the ST405 clone was found in six of the eight hospitals with OXA-48 carbapenemase.
The expansion of all these clones is taking place in the elderly population and is related to the healthcare system. These results are similar to those obtained from patients infected by ESBL- or acAmpC-producing strains.27
Like ST405, the sequence types ST101, ST14 and ST17 are also described as multiresistant clones present in various European and American countries.7,28–31 Nevertheless, in our case, ST14 and ST17 were only detected exceptionally and did not carry additional resistances. Finally, ST1233, described here for the first time, was a single locus variant of ST540, and did not show resistance to aminoglycosides or quinolones. All these clones were isolated in a timely manner.
Taken together, our results show that the increasing prevalence of carbapenemase OXA-48 in Catalonia is due to the expansion of the K. pneumoniae ST405 clone. All clones described preserved the blaOXA-48 genetic environment as well as the mobile genetic elements (Tn1999). Curiously, the three strains with minority ST were not resistant to multiple drugs, perhaps because of an absence of selection pressure due to their infrequency and therefore low exposure to antibiotics.
Conflict of interestThere is no conflict of interest.
We thank all participant health centres for their willingness to take part in the study and also the group of microbiologists of county hospitals in Catalonia and the Balearic Islands (http://www.scmimc.org/grupstreball02.php) for collecting the strains and epidemiological data.