Fluoroquinolone resistance in methicillin-resistant Staphylococcus aureus (MRSA) has increased in recent years. The objective of this study was to characterise two MRSA populations, one susceptible to fluoroquinolones and other resistant identifying the clonal types and the differential characteristics of both MRSA populations.
MethodsMolecular typing using PFGE, MLST, spa and SSCmec was performed on 192 MRSA strains isolated from 2009 to 2011, 49 only oxacillin-resistant (OX-R) and 143 oxacillin and levofloxacin-resistant (OX-R-LEV-R). Mutations that conferred resistance to fluoroquinolones, hypermutable phenotypes and the presence of eight microbial surface components recognising adhesive matrix molecules (MSCRAMMs) were also studied.
ResultsA statistically significant increase in the OX-R-LEV-R phenotype was observed (p<0.05). The most common clone of the OX-R isolates was sequence type (ST) 8 (32.6%), followed by ST72 (26.5%) and ST5 (26.5%). In the OX-R-LEV-R phenotype, the ST5 clone was the most common (65.7%), followed by ST72 (15.4%), and ST125 (12.6%). All isolates except the ST398 clone carried the SCCmecIVc. Clones ST5, ST72, ST125, and ST30 had hypermutable phenotypes. The ST72 clone and the ST30 clone in the OX-R phenotype harboured the highest number of MSCRAMMs.
ConclusionST5 and ST72 clones were the most frequent clones identified in OX-R-LEV-R phenotype. Both clones showed a hypermutable phenotype that favours their selection as the fluoroquinolone resistant clones. The genetic relationships identified indicate that OX-R-LEV-R clones have evolved from OX-R MRSA clones.
La resistencia a fluoroquinolonas en Staphylococcus aureus resistente a meticilina (SARM) se ha incrementado en los últimos años. El objetivo de este estudio consistió en caracterizar 2 poblaciones de SARM, una sensible a fluoroquinolonas y otra resistente identificando los tipos clonales y las características diferenciales entre los mismos.
MétodosEn un total de 192 SARM aislados entre los años 2009-2011, 49 solo oxacilina resistentes (OX-R) y 143 oxacilina y levofloxacino resistentes (OX-R-LEV-R), se realizó el tipado molecular mediante PFGE, MLST, spa y SSCmec. Además se estudiaron las mutaciones que confieren resistencia a las fluoroquinolonas, los fenotipos hipermutadores y la presencia de 8 componentes de la superficie microbiana que reconocen adhesinas de la matriz extracelular.
ResultadosEn el periodo de estudio se detectó un incremento estadísticamente significativo del fenotipo OX-R-LEV-R (p<0,05). Entre los OX-R el clon ST8 (32,6%) fue el más frecuente seguido de los clones ST72 (26,5%) y ST5 (26,5%). Entre los aislados del fenotipo OX-R-LEV-R, el clon ST5 fue el más frecuente (65,7%), seguido de los clones ST72 (15,4%) y ST125 (12,6%). Todos los aislamientos, excepto el clon ST398, portaban el SCCmec-IVc. Los clones ST5, ST30, ST72 y ST125 presentaron un fenotipo hipermutador. Los clones ST72 y ST30 OX-R son los que poseen una mayor dotación de componentes de la superficie microbiana que reconocen adhesinas de la matriz extracelular.
ConclusiónLos clones ST5 y ST72 fueron los más frecuentes en el fenotipo OX-R-LEV-R. Ambos clones poseían un fenotipo hipermutador. La estrecha relación genética entre los clones OX-R y OX-R-LEV-R pertenecientes al mismo ST sugiere que estos últimos han evolucionado a partir de una población OX-R preexistente.
Methicillin-resistant Staphylococcus aureus (MRSA) causes a large number of infections.1 Its ability to adapt and acquire resistance to different antimicrobials has possibly favoured its spread both in hospitals and in the community.1 Methicillin resistance in S. aureus is coded by the mec gene, included in a genomic island called the staphylococcal cassette chromosome mec (SCCmec). The fluoroquinolones are among the therapeutic agents whose activity is not affected by SCCmec. However, rates of fluoroquinolone resistance have increased,2 mainly due to mutations in genes gyrA and grlA.2
S. aureus expresses surface proteins that are essential for its success both as a commensal and as a pathogen. The most important group of these proteins are the MSCRAMMs (Microbial Surface Components Recognizing Adhesive Matrix Molecules).3
The objective of this study was to analyse the increase in MRSA resistant to fluoroquinolones and its possible relation with a clonal selection. In addition, the characteristics of the MRSA isolates studied were analysed including the phenotypic hypermutability of the identified clones and the MSCRAMMs content.
Materials and methodsSelection of microorganisms and patients for inclusion in the studyThe University Hospital Complex of Pontevedra has a catchment population of 296,463. We selected two groups of MRSA isolates from the years 2009 to 2011: (i) those that presented resistance only to beta-lactams (oxacillin, OX-R), being susceptible to all other groups of antimicrobials; and (ii) those with resistance to oxacillin and to levofloxacin OX-R-LEV-R, being susceptible to all other antimicrobial groups. Only one isolate per patient was analysed in the study. A total of 192 isolates were studied. The following patient data were recorded: sex, age, type of sample, underlying diseases and risk factors for MRSA infection. The criteria of the Centers for Disease Control and Prevention (CDC) were applied to determine the origin of the MRSA infection.4,5
Bacterial identification and susceptibility to antimicrobialsThe biochemical characteristics and antibiotic susceptibility of the isolates were studied using the commercially available Wider system (Gram Positive MIC/ID Panel, Francisco Soria Melguizo, S.A.). The minimum inhibitory concentrations (MICs) for fluoroquinolones were determined using the gradient diffusion (epsilon-test) method. Resistance to methicillin was detected using a 30μg cefoxitin disc. The European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines were followed for the determination of breakpoints and resistance to methicillin.6
Molecular typing of the isolatesFor the SCCmec, PFGE, and spa gene typing, the 192 strains included in the study were analysed.
Analysis of the staphylococcal cassette chromosome mec (SCCmec): a multiplex polymerase chain reaction (PCR) was employed to identify the mecA gene and SCCmec7 type and subtype.
Pulsed-field gel electrophoresis (PFGE): cells were processed following the protocol proposed by Murchan et al.8 Gels were analysed using the GelCompar II software (Applied Maths NV), applying the Dice similarity coefficient with an optimisation of 0.5% and tolerance 1%. Banding patterns were assigned to pulsotypes (using uppercase Latin letters) and subtypes (using numerical subindices). Pulsotypes were considered to be different if the coefficient of similarity was less than 80%. Subtypes were defined as pulsotypes with a coefficient of similarity between 80% and 95%.9
spa gene typing: amplification and sequencing of the X region of the spa gene were performed under the conditions described by Harmsen et al.10 The spa type was established using Ridom StaphType software (Ridom GmbH, Würzburg, Germany).
Multilocus sequence typing (MLST): the Enright et al.11 recommendations were followed. A total of 17 strains were studied including one isolate from each of the identified pulsotypes and spa types. The MLSTs of the strains not analysed by this method were deduced from the spa types.
Analysis of the frequency of mutation in the isolatesThe frequency of mutations for rifampicin and streptomycin were performed under the conditions described by Trong et al.12 One isolate from each clonal type and one from each phenotype were selected and processed in triplicate. In summary, five OX-R strains including one of each ST5, ST72, ST30, ST8, and ST398, and another five OX-R-LEV-R strains including one of each ST5, ST72, ST30, ST125, and ST22 were studied.
Analysis of mutations in genes coding for fluoroquinolone resistanceThe amplification of genes gyrA and grlA was carried out under the conditions described by Schmitz et al.13 A total of 30 strains belonging to the ST5 clone, 22 strains belonging to the ST72, 18 strains from the ST125, 8 strains from the ST22, and 1 strain from the ST30 were studied. Sequence analysis was performed using the BioEdit software, version 7.1.9 (Ibis Therapeutics, Isis Pharmaceuticals, Inc.).
Detection of genes coding for surface proteinsBased on the relationship between the surface proteins and the S. aureus colonisation, we selected eight genes that coded for surface proteins belonging to the MSCRAMM group3: clfA,14clfB,14fnbA,15fnbB,16cna,15sdrC,14ebpS14 and efb.15 The presence of these genes was determined by PCR under the conditions described by the authors cited for each MSCRAMM studied. The 192 strains included in the study were analysed.
Statistical analysisThe chi square test for trend was used to analyse the years of the study and the OX-R-LEV-R increase. To analyse the mutation rate to streptomycin and rifampicin, the non-parametric test of Mann–Whitney was used to compare two groups of the studied strains. The non-parametric Kruskal–Wallis test was used for multiple comparisons. A p value less than or equal to 0.05 was considered significant. Statistical analysis was performed using the statistical package SPSS 22.0.
ResultsCharacteristics of the isolates and molecular typing of MRSABetween 2009 and 2011, a total of 2587 S. aureus were isolated; 728 (28.1%) were MRSA. Of the 728 MRSA isolates, 49 (6.7%) were only resistant to beta-lactams (OX-R) and 143 (19.6%) were OX-R-LEV-R (Table 1). Throughout the three years of the study (2009–2011), an increase of 9.1% was identified in OX-R-LEV-R MRSA being statistically significant (p<0.05), whereas OX-R MRSA isolates only increased by 2.1% (Table 1). A total of 60 MRSA were considered community-associated MRSA (CA-MRSA) strains (31.3%); this community origin was found in 44.9% (n=22) of the OX-R phenotype and in 26.6% (n=38) of the OX-R-LEV-R phenotype.
Staphylococcus aureus isolated between 2009 and 2011 at the University Hospital Complex of Pontevedra (%).
| Year | S. aureus (no. isolates) | Total MRSA | Only (OX)-R MRSA | p-values | (OX-LEV)-R MRSA | p-values |
|---|---|---|---|---|---|---|
| 2009 | 821 | 229 (27.9) | 13 (5.7) | 34 (14.9) | ||
| 2010 | 864 | 241 (27.9) | 16 (6.6) | 0.361 | 47 (19.5) | 0.011 |
| 2011 | 902 | 258 (28.6) | 20 (7.8) | 62 (24) |
Abbreviations: LEV, levofloxacin; MRSA, methicillin-resistant Staphylococcus aureus; OX, oxacillin; R, resistant.
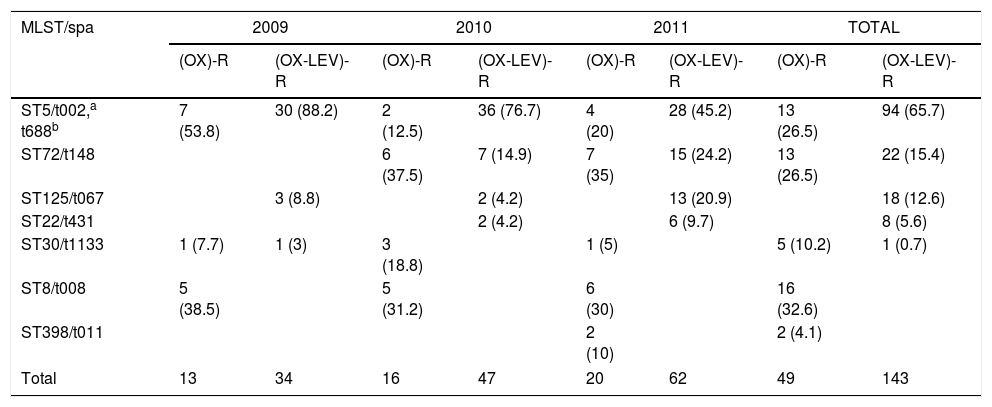
Ten pulsotypes (A–J) were identified (Fig. 1). The following subtypes were identified: four in pulsotype A, three in pulsotype E, and two in pulsotype G (Fig. 1). The spa and ST identified in the OX-R and OX-R-LEV-R phenotypes are shown in Table 2. All isolates carried a SCCmecIVc, except ST398 that harboured SCCmecV.
MRSA clonal types identified in the study.
*ST30 clone showed the same PFGE profile (PFGE-J) in (OX)-R and (OX-LEV)-R phenotypes. Abbreviations: LEV, levofloxacin; MRSA, methicillin-resistant Staphylococcus aureus; OX, oxacillin; PFGE, pulsed-field gel electrophoresis; R, resistant; ST, sequence type.
Yearly variation of clonal types, SARM (OX)-R and (OX-LEV)-R (%).
| MLST/spa | 2009 | 2010 | 2011 | TOTAL | ||||
|---|---|---|---|---|---|---|---|---|
| (OX)-R | (OX-LEV)-R | (OX)-R | (OX-LEV)-R | (OX)-R | (OX-LEV)-R | (OX)-R | (OX-LEV)-R | |
| ST5/t002,a t688b | 7 (53.8) | 30 (88.2) | 2 (12.5) | 36 (76.7) | 4 (20) | 28 (45.2) | 13 (26.5) | 94 (65.7) |
| ST72/t148 | 6 (37.5) | 7 (14.9) | 7 (35) | 15 (24.2) | 13 (26.5) | 22 (15.4) | ||
| ST125/t067 | 3 (8.8) | 2 (4.2) | 13 (20.9) | 18 (12.6) | ||||
| ST22/t431 | 2 (4.2) | 6 (9.7) | 8 (5.6) | |||||
| ST30/t1133 | 1 (7.7) | 1 (3) | 3 (18.8) | 1 (5) | 5 (10.2) | 1 (0.7) | ||
| ST8/t008 | 5 (38.5) | 5 (31.2) | 6 (30) | 16 (32.6) | ||||
| ST398/t011 | 2 (10) | 2 (4.1) | ||||||
| Total | 13 | 34 | 16 | 47 | 20 | 62 | 49 | 143 |
The yearly increase in the OX-R-LEV-R phenotype is shown in Table 2. The prevalence of ST5 decreased throughout the three years, from 88.2% in 2009 to 45.2% in 2011. Nevertheless, ST72 increased its prevalence from zero isolates in 2009 to 24.2% in 2011. In addition, ST125 increased its prevalence from 8.8% in 2009 to 20.9% in 2011.
Characteristics of the population studiedThe 192 MRSA isolates corresponded to 192 patients. The median age of the patients was 77 years (range 10 days–94 years), and 50.0% were men. The most common underlying diseases associated with isolation of MRSA were cardiovascular diseases (54.2%), followed by diabetes mellitus (35.8%) and cancer (14.7%); two or more concurrent underlying diseases were identified in 30.3% of patients. In 28.4% of patients there was no detectable underlying disease. The source of the isolates was skin and soft tissue infection in 71.7% of cases, followed by bacteraemia (6.6%). Factors related to MRSA infection included hospitalisation in the 12 months prior to the isolation of the microorganism (42.1%) and a previous history of MRSA infection (19.2%).
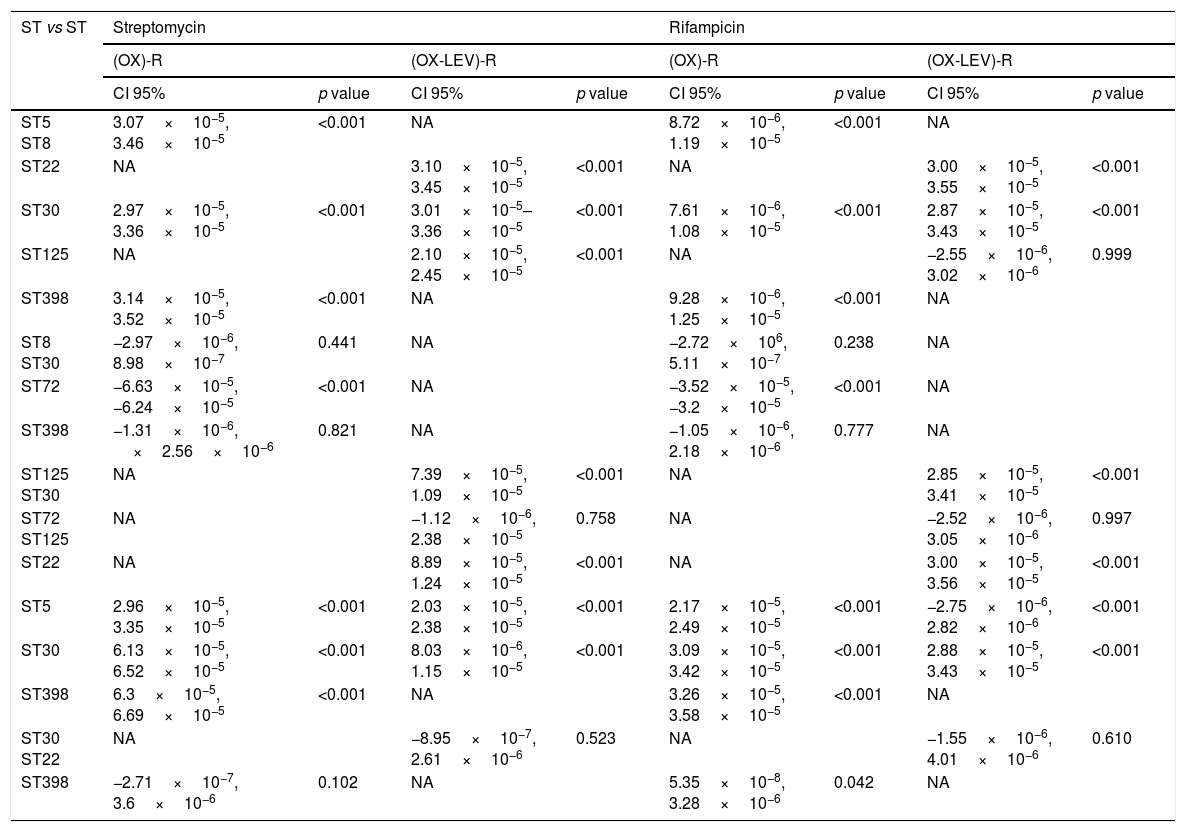
Fluoroquinolone mutations and clonal hypermutants identified in OX-R and OX-R-LEV-R MRSAAll the OX-R-LEV-R strains showed a levofloxacin MIC of >32mg/l and the mutations S84L, in gyrA, and S80F in grlA. In addition to mutation S84L, the isolates belonging to clone ST22 carried the mutation S85P in gyrA. Mutation frequency experiments revealed a higher frequency of mutations (10−5–10−6) for clonal types ST5, ST30, ST72, and ST125 compared with clonal types ST8, ST22 and ST398 (10−8–10−7). No statistically differences in mutation rates were detected when comparing the OX-R strains, or the OX-R-LEV-R strains. The mutation rate when comparing clones was statistically significant in the OX-R and OX-R-LEV-R phenotypes (p<0.05) both for streptomycin and rifampicin. Analysing the mutation rate of the different pairs of clones (Table 3), the differences were statistically significant for all clones except ST8 vs ST30, ST8 vs ST398, and ST30 vs ST398 in the OX-R phenotype both for streptomycin and rifampicin. The mutation rate for streptomycin of the OX-R-LEV-R phenotype was statistically significant in all pair of clones except ST30 vs ST22 and ST72 vs ST125 (Table 3). The mutation rate for rifampicin of the OX-R-LEV-R phenotype was statistically significant in all pair of clones except ST5 vs ST125, ST72 vs ST125, and ST30 vs ST22 (Table 3).
Multiple comparison in the mutation rate for SARM (OX)-R and (OX-LEV)-R phenotypes.
| ST vs ST | Streptomycin | Rifampicin | ||||||
|---|---|---|---|---|---|---|---|---|
| (OX)-R | (OX-LEV)-R | (OX)-R | (OX-LEV)-R | |||||
| CI 95% | p value | CI 95% | p value | CI 95% | p value | CI 95% | p value | |
| ST5 ST8 | 3.07×10−5, 3.46×10−5 | <0.001 | NA | 8.72×10−6, 1.19×10−5 | <0.001 | NA | ||
| ST22 | NA | 3.10×10−5, 3.45×10−5 | <0.001 | NA | 3.00×10−5, 3.55×10−5 | <0.001 | ||
| ST30 | 2.97×10−5, 3.36×10−5 | <0.001 | 3.01×10−5–3.36×10−5 | <0.001 | 7.61×10−6, 1.08×10−5 | <0.001 | 2.87×10−5, 3.43×10−5 | <0.001 |
| ST125 | NA | 2.10×10−5, 2.45×10−5 | <0.001 | NA | −2.55×10−6, 3.02×10−6 | 0.999 | ||
| ST398 | 3.14×10−5, 3.52×10−5 | <0.001 | NA | 9.28×10−6, 1.25×10−5 | <0.001 | NA | ||
| ST8 ST30 | −2.97×10−6, 8.98×10−7 | 0.441 | NA | −2.72×106, 5.11×10−7 | 0.238 | NA | ||
| ST72 | −6.63×10−5, −6.24×10−5 | <0.001 | NA | −3.52×10−5, −3.2×10−5 | <0.001 | NA | ||
| ST398 | −1.31×10−6, ×2.56×10−6 | 0.821 | NA | −1.05×10−6, 2.18×10−6 | 0.777 | NA | ||
| ST125 ST30 | NA | 7.39×10−5, 1.09×10−5 | <0.001 | NA | 2.85×10−5, 3.41×10−5 | <0.001 | ||
| ST72 ST125 | NA | −1.12×10−6, 2.38×10−5 | 0.758 | NA | −2.52×10−6, 3.05×10−6 | 0.997 | ||
| ST22 | NA | 8.89×10−5, 1.24×10−5 | <0.001 | NA | 3.00×10−5, 3.56×10−5 | <0.001 | ||
| ST5 | 2.96×10−5, 3.35×10−5 | <0.001 | 2.03×10−5, 2.38×10−5 | <0.001 | 2.17×10−5, 2.49×10−5 | <0.001 | −2.75×10−6, 2.82×10−6 | <0.001 |
| ST30 | 6.13×10−5, 6.52×10−5 | <0.001 | 8.03×10−6, 1.15×10−5 | <0.001 | 3.09×10−5, 3.42×10−5 | <0.001 | 2.88×10−5, 3.43×10−5 | <0.001 |
| ST398 | 6.3×10−5, 6.69×10−5 | <0.001 | NA | 3.26×10−5, 3.58×10−5 | <0.001 | NA | ||
| ST30 ST22 | NA | −8.95×10−7, 2.61×10−6 | 0.523 | NA | −1.55×10−6, 4.01×10−6 | 0.610 | ||
| ST398 | −2.71×10−7, 3.6×10−6 | 0.102 | NA | 5.35×10−8, 3.28×10−6 | 0.042 | NA | ||
NA: not applicable.
Except for ST30, any single clonal type possessed certain specific MSCRAMMs independently of the resistance phenotype. The following MSCRAMMs were detected: ST72-(clfA-clfB-fnbA-fnbB-sdrC-efb), ST30-OX-R-(clfA-clfB-fnbA-cna-sdrC-ebpS), ST30-OX-R-LEV-R-(clfA-clfB-fnbA-sdrC-ebpS), ST5-(clfA-clfB-fnbA-fnbB-ebpS), ST8-(clfA-clfB-fnbA-sdrC-efb), ST22-(clfA-clfB-fnbA-cna), ST125-(clfA-clfB-fnbA-ebpS), ST398-(clfA-fnbA).
DiscussionMRSA infections are more common in older individuals with comorbidities.17 In our study, CA-MRSA was the cause of 31.3% of the infections. This figure is higher than that previously reported in several nationwide studies in Spain18 and in Europe.19 However, the rates in the United States are much higher, above 50%.20
The increase in the prevalence of resistance to fluoroquinolones has been associated with different factors21 including the persistent increase in quinolone prescriptions, as is the case of Spain.22 The main mechanism of resistance to fluoroquinolones is through mutations in the genes coding for topoisomerase IV and DNA gyrase, and the degree of resistance is proportional to the number of mutations accumulated.23
The international epidemic clone ST5 was the most successful in our study (Table 2). Over the first decade of this century, this clone has persisted in our healthcare setting carrying SCCmecII.24 It currently carries SCCmecIVc, and is at present the main clone isolated.9,25 The smaller size of SCCmecIVc may be an evolutionary advantage, making its spread more effective.26 The fact that this clone circulated for a longer period in our setting,24 and its hypermutable status may facilitate its selection as the predominant clone of the fluoroquinolone-resistant phenotype.
ST72 is also a hypermutable clone that emerged recently in our setting, and a significant increase in its prevalence has been reported.25 Although this clone is uncommon in Europe, we must remain alert to its probable spread; in South Korea it is the primary cause of community-acquired MRSA infection, and it is also the main MRSA isolated from the meat of animals for consumption.27 Its high number of MSCRAMMs may be related to its elevated capacity to colonise both humans and animals.
The close clonal relationship between most of the OX-R and OX-R-LEV-R strains in ST5, ST72, and ST30 clones suggests that quinolone resistance evolved from the OX-R population. Whereas ST30 is infrequent in our study, ST5 and ST72 were the two most common clones isolated. Interestingly, these were the only clones in which fnbB-fnbA were detected simultaneously. These two genes code for adhesins important in the adhesion and internalisation of S. aureus into mammalian epithelial cells and in the initiation of the biofilm.3 This may have contributed to their epidemiological expansion.
The pandemic clone ST30 is highly prevalent in the Southwest Pacific, but is infrequent in Spain,18 representing only 3.1% of the isolates in our study.
The international epidemic clones ST22 and ST125 have been isolated frequently in Spain, mainly clone ST125.9,28 In our study, only strains belonging to the OX-R-LEV-R phenotype were detected belonging to these clones. Since no OX-R isolates were detected, we can speculate that these two clones were introduced in the MRSA population as OX-R-LEV-R and not evolved from a previous OX-R local population.
The ST8 clone has been selected as the major clonal type in the OX-R phenotype (32.6%). However, we did not identify ST8 OX-R-LEV-R strains in the present study, a finding that could be related to the fact that it is not a hypermutable clone.
The ST398 clone had the smallest number of MSCRAMMs and lacked the two adhesins (clfB-sdrC) with affinity for epithelial cells. This fact could limit its ability to colonise the nares in humans as ST398 is a clone typically associated with livestock.29
The ST5 and ST72 clones were the most frequently identified in the OX-R-LEV-R population. Both clones showed a hypermutable phenotype that favours their selection as the fluoroquinolone resistant clones. The genetic relationships identified between OX-R and OX-R-LEV-R clones that belong to the same ST indicate that OX-R MRSA clones have evolved from a pre-existing OX-R MRSA population. The richest MSCRAMMs environment in the most frequent clones could have played a role in their fitness to colonise the host.
FundingThis work was partially financed by Fondo de Investigaciones Sanitarias PI070812 and Consejería de Sanidad de la Junta de Galicia PS08/34.
Conflict of interestThe authors declare that they have no conflict of interest.
We would like to thank Dr. Marta García Campello, Head of the Microbiology and Parasitology Laboratories Department of the University Hospital Complex of Pontevedra, for allowing us access to the strains under study. We would also like to thank Dr. José Pintado Valverde of Marinas de Vigo Research Institute (CSIC) for his collaboration in the running of the GelCompar II software. We would also like to thank Dr. Ángel Salgado Barreira for the statistical analysis.