Molecular and genomic pathology is an essential cornerstone of diagnosis in breast disease, to such an extent that genetic information is already included in therapeutic decision-making. There are now various commercial platforms available in the clinic, generally with little or no agreement in the genes included, in their technical basis, in the definition of risk groups, in the information they provide, in their indications or in the protocols required to use them.
ObjectiveTo evaluate the use and knowledge of these platforms.
Material and methodsAn eleven-question survey was conducted targeting breast units accredited by the SESPM in Spain at that time.
Results26 units out of the 36 surveyed responded and data was obtained that can guide the use of the platforms and serve as a starting point towards gaining a deeper knowledge of them.
ConclusionsThe indications approved by the Autonomous Regions need to be re-evaluated. There is insufficient evidence to base decisions about the axilla on the platforms. MammaPrint® is the only platform with level of evidence 1a for N1–3 patients. It also identifies a subgroup of patients who may not require hormonal treatment.
La enfermedad molecular y genómica constituye un pilar irrenunciable del diagnóstico en enfermedad mamaria, de tal manera que la información genética ha sido ya integrada en la toma de decisiones terapéuticas. Actualmente, existen diferentes plataformas comerciales disponibles en la clínica, generalmente con pocas o nulas coincidencias en los genes incluidos, en su fundamento técnico, en la definición de grupos de riesgo, en la información que proporcionan, en sus indicaciones y en el circuito requerido para la realización de las mismas.
ObjetivoEvaluar el uso y conocimiento de dichas plataformas.
Material y métodosSe realizó una encuesta de 11 preguntas dirigidas a las unidades de mama acreditadas en España por la SESPM en ese momento.
ResultadosRespondieron 26 unidades de 36 encuestadas y se obtuvieron datos que pueden ser orientativos acerca del uso de las plataformas y pueden servir como punto de partida para profundizar en su conocimiento.
ConclusionesEs necesario re-evaluar indicaciones aprobadas por las CC.AA. No existe evidencia suficiente para tomar decisiones sobre la axila en función de la plataforma. MammaPrint® es la única plataforma con evidencia IA para pacientes N1-3. Además, identifica un subgrupo de pacientes que pueden no requerir tratamiento hormonal.
Breast cancer is a heterogeneous disease not only from a morphological perspective but also from a genetic perspective, with highly variable disease-free and relapse-free intervals that can range from months to decades from the time of initial diagnosis and treatment.
Therapeutic decision-making is based, fundamentally, on the characteristics of the tumour, both morphological and biological, and on its extent. Recently, genetic information has also been integrated to identify patients with high clinical-pathological risk (HR), but low genomic risk (LR), who do not require chemotherapy.
Given that molecular and genomic pathology already constitute an essential pillar of diagnosis in breast pathology, multiple commercial platforms are available in the clinic, generally with few or no coincidences in the genes included, in their technical basis, in the definition of risk groups, in the information they provide, in their indications and in the circuit required to carry them out.
ObjectivesTo explore the degree of knowledge we have of genomic platforms for breast cancer in daily practice, from the SESPM and through a brief survey directed at the 36 SESPM-accredited Breast Units at that time. The survey was conducted in June 2019 and consisted of 11 questions with a dichotomous (yes/no) or multiple response depending on the type of question.
To comment on this work, from these questions we selected those that have seemed to be of greatest interest to us, either because of the question itself or because of the answers obtained.
Twenty-six units answered the questionnaire, i.e., 72.2% of those surveyed. By specialty, there were 10 gynaecologists, 5 surgeons, 9 oncologists, and 2 pathologists.
Material and methodsThe questions and possible answers were made through the SurveyMonkey platform to facilitate the online response of the respondents, as well as the subsequent analysis of the data obtained.
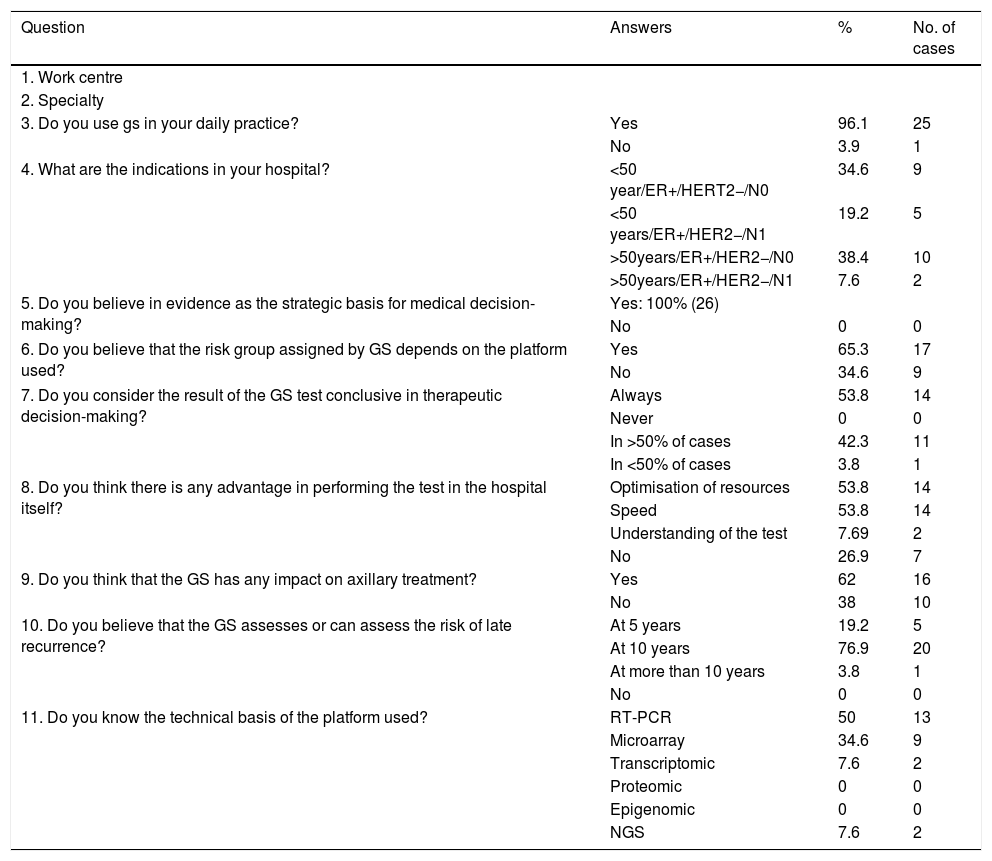
Table 1 shows the questions asked, the possible answers and the percentages broken down for each answer.
| Question | Answers | % | No. of cases |
|---|---|---|---|
| 1. Work centre | |||
| 2. Specialty | |||
| 3. Do you use gs in your daily practice? | Yes | 96.1 | 25 |
| No | 3.9 | 1 | |
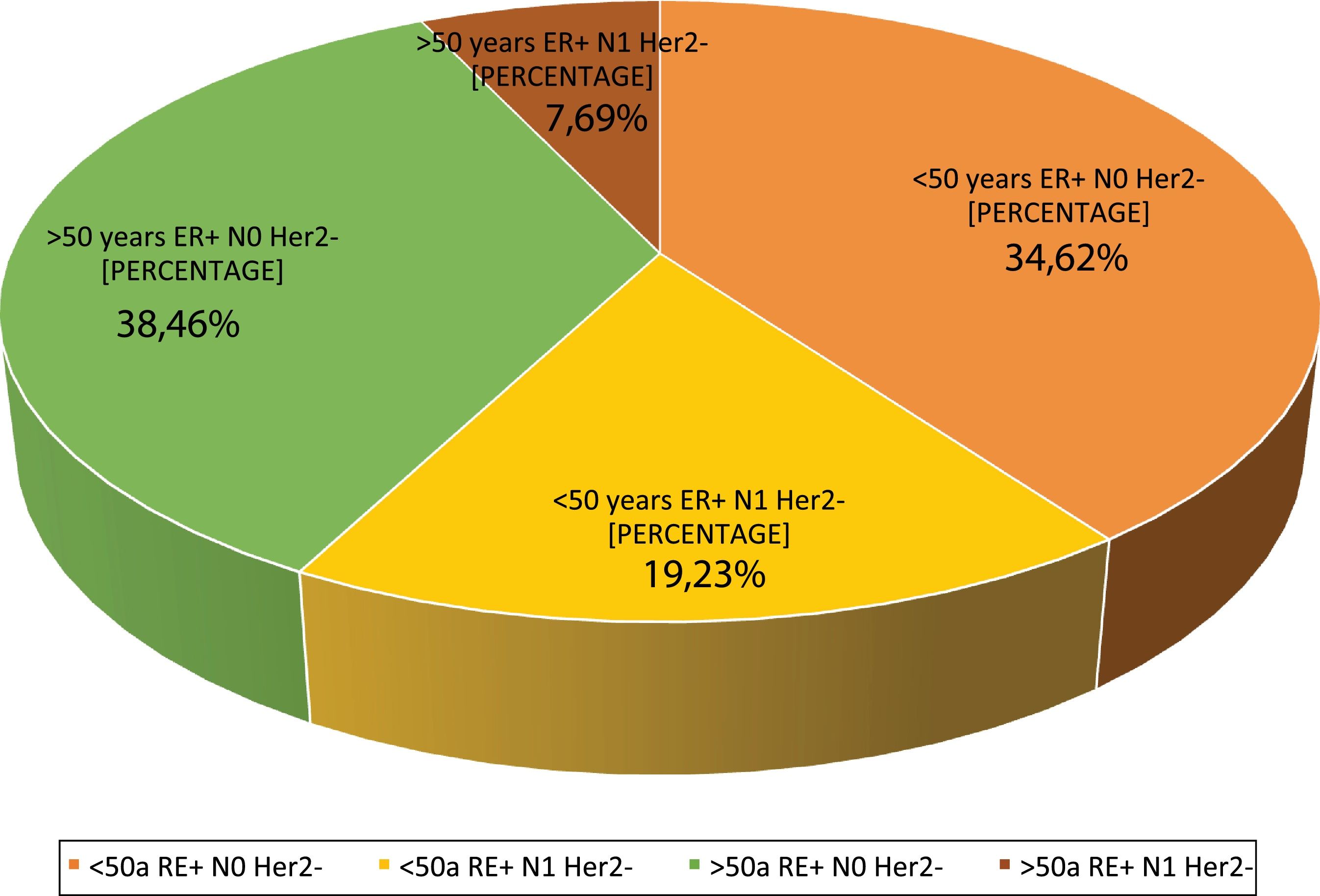
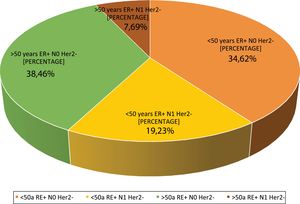
| 4. What are the indications in your hospital? | <50 year/ER+/HERT2−/N0 | 34.6 | 9 |
| <50 years/ER+/HER2−/N1 | 19.2 | 5 | |
| >50years/ER+/HER2−/N0 | 38.4 | 10 | |
| >50years/ER+/HER2−/N1 | 7.6 | 2 | |
| 5. Do you believe in evidence as the strategic basis for medical decision-making? | Yes: 100% (26) | ||
| No | 0 | 0 | |
| 6. Do you believe that the risk group assigned by GS depends on the platform used? | Yes | 65.3 | 17 |
| No | 34.6 | 9 | |
| 7. Do you consider the result of the GS test conclusive in therapeutic decision-making? | Always | 53.8 | 14 |
| Never | 0 | 0 | |
| In >50% of cases | 42.3 | 11 | |
| In <50% of cases | 3.8 | 1 | |
| 8. Do you think there is any advantage in performing the test in the hospital itself? | Optimisation of resources | 53.8 | 14 |
| Speed | 53.8 | 14 | |
| Understanding of the test | 7.69 | 2 | |
| No | 26.9 | 7 | |
| 9. Do you think that the GS has any impact on axillary treatment? | Yes | 62 | 16 |
| No | 38 | 10 | |
| 10. Do you believe that the GS assesses or can assess the risk of late recurrence? | At 5 years | 19.2 | 5 |
| At 10 years | 76.9 | 20 | |
| At more than 10 years | 3.8 | 1 | |
| No | 0 | 0 | |
| 11. Do you know the technical basis of the platform used? | RT-PCR | 50 | 13 |
| Microarray | 34.6 | 9 | |
| Transcriptomic | 7.6 | 2 | |
| Proteomic | 0 | 0 | |
| Epigenomic | 0 | 0 | |
| NGS | 7.6 | 2 |
The results were presented and discussed at the 4th Spanish Breast Congress (4th CEMA). For publication, the authors selected, at their discretion, the eight questions of greatest interest in light of the answers, referenced in the text according to their numbering in Table 1 and whose comments are based on the review of the literature.
Our objectives were in no case to make judgments based on the respondents’ answers, but rather to obtain the real picture of the use of the platforms in our environment, providing, where we thought appropriate, relevant information that, in our opinion, might be of interest.
ResultsQuestion 3: Do you use genomic signature (GS) in your daily practice?96.1% of respondents answered affirmatively. Therefore, almost 4% of respondents do not use any genomic platform in their daily practice. Given that the use of genomic platforms is financed in our country for specific indications and that their use is recommended in all clinical guidelines, SESPM, NCCN, NICE, ASCO, EGTM, the St. Gallen panel of experts and even in the latest edition of the TNM, it is worth considering whether the reasons for not applying them could be due to the logistical difficulty in handling the tissue, distrust in the results, or because it is considered that, for therapeutic decision-making, the immunohistochemical study is sufficient. The fact that the new extension of the indications (patients with 1–3 positive lymph nodes), is not financed by some autonomous communities, may be another limitation to its use.
Question 4: What are the indications at your hospital? This question had four possible answers: <50 years, RE+, HER2−, N0, <50 years, RE+, HER2−, N1, >50 years, RE+, HER2−, N0 and >50 years, RE+, HER2−, N1.For most respondents, GS is primarily used in RE+/HER2−/N0 patients, regardless of their age (Fig. 1). Among the respondents, therefore, age is not a variable that affects the use of GS. Although age is not observed to be a limiting condition for use of the platform, we should remember that only MammaPrint®, Oncotype Dx® and EndoPredict® are approved for both pre- and post-menopausal patients.
Regarding the use of GS in N1 patients, only a quarter of the respondents (27%) use it in this subgroup of patients, while 73% only use it in RE+/HER2−/N0 patients. Although currently available evidence supports its use in 1–3 positive lymph nodes, the lack of funding from some autonomous communities for N1 cases is a major constraint to its use in that clinical setting.
Currently, MammaPrint® is the only GS with a 1A evidence level for use in N1-3 patients. The evidence level for Oncotype Dx®, Prosigna® and EndoPredict® in these patients is currently only 2B (NICE Guide 2020).1–4
Question 5: Do you believe in evidence as a strategic basis for medical decision-making?100% of the respondents answered affirmatively. The meaning of the evidence and degrees of recommendation, despite the value given to that condition by most respondents, is not always clear. Levels of evidence are based on study design and methodological quality of individual studies. The degrees of recommendation are based on the strength of the evidence supporting the study. The hierarchy of the different types of studies is from more to less, systematic reviews and meta-analyses of controlled and randomised trials, observational studies, non-experimental studies, and expert opinion. The highest levels of evidence are acquired on the basis of high quality meta-analyses and systematic reviews while the lowest levels of evidence are based on expert opinion.
There are several evidence classification models, but the one generally used is that proposed by the Centre for Evidence-Based Medicine, Oxford (OCEBM)5 that classifies evidence into levels ranging from 1 to 5, (with level 1 being the “best evidence” and level 5 being “worse”, “the worst” or “the least good”), together with recommendation grades A–D. This model has the advantage of assuring us of the most accurate knowledge of each scenario, owing to its high degree of specialisation. Furthermore, it has the prerogative to clarify how a lack of methodological rigour affects the design of the studies, diminishing their value not only in the gradation of evidence, but also in the strength of recommendations. In Table 2, we recall the differences between levels of evidence 1.
Based on the Oxford classification of levels of evidence (OCEBM).5
| Grade of recommendation | Level of evidence | Treatment, prevention, aetiology and damage | Prognosis and natural history | Diagnosis | Differential diagnosis and prevalence study | Economic studies and decision analysis |
|---|---|---|---|---|---|---|
| A | 1a | SR with homogeneity of controlled CT with random assignment | SR of cohort studies, with homogeneity, i.e., that includes studies with comparable results, in the same direction and validated in different populations | SR of diagnostic studies of level 1 (high quality), with homogeneity, i.e., that includes studies with comparable results and in the same direction and in different clinical centres | SR with homogeneity of prospective cohort studies | SR with homogeneity of level 1 economic studies |
| 1b | Individual CT with narrow confidence interval | Individual cohort studies with follow-up of more than 80% of the cohort and in a single population | Cohort studies that validate the quality of a specific test, with an appropriate reference standard (independent of the test) or from algorithms for estimation of prognosis or classification of diagnosis or tested in a clinical centre | Prospective cohort study with good follow-up | Analysis based on cost or clinically sensitive alternatives; SR of the evidence; and including sensitivity analysis | |
| 1c | Efficiency demonstrated by clinical practice. Considers whether some patients die before they are assessed | Results based on effectiveness and not on their efficacy demonstrated through a cohort study. All or none case series | Diagnostic tests with specificity so high that a positive result confirms the diagnosis and with sensitivity so high that a negative result rules out the diagnosis | All or none case series | Absolute analysis in terms of the highest or lowest value | |
| B | 2a | SR of cohort studies, with homogeneity | SR of retrospective cohort studies or untreated control groups in a CT, with homogeneity | Sr of level 2 diagnostic studies (medium quality) with homogeneity | SR (with homogeneity) of 2b studies and better | SR (with homogeneity) of economic studies with level higher than 2 |
| 2b | Individual cohort study with less than 80% follow-up (includes low-quality CT) | Retrospective cohort study or follow-up of untreated controls in a CT or non-validated CPG | Exploratory studies which, through logistic regression, determine significant factors and are validated with an appropriate reference standard (independent of the test) | Retrospective or insufficient follow-up cohort studies | Analysis based on costs or clinically sensitive alternatives; limited to review of the evidence; and including a sensitivity analysis |
Systematic reviews (SR), randomised controlled clinical trials (CT), clinical practice guidelines (CPG).
It is worth noting that, although 100% of respondents believe in evidence as the basis for medical decision-making, they use tests with a 2A evidence level despite the fact that tests with a 1A evidence level are available on the market.
Question 6: Do you think the risk group assigned by the genomic signature depends on the platform used?65.3% (17 answers) of the respondents believed that it did and 34.6% (9 answers) believed that it did not. In recent decades, the recognition of different intrinsic molecular subtypes has highlighted the complexity of breast cancer biology. Although immunohistochemistry (IHC) is the most commonly applied technique for the surrogate molecular classification of breast cancer, gene-expression-based assays (BluePrint®, PAM50) evaluate a greater number of genes than IHC/FISH surrogates and can reclassify a significant number of patients based on molecular pathways. This reclassification can have important consequences for therapeutic decision-making, tumour response to specific drugs and clinical follow-up. Consequently, two fundamental questions arise:
- 1.
Is there concordance between the intrinsic subtype assigned by genomic platform and the intrinsic-like subtype assigned by IHC? If not, does the genomic platform add information to the IHC subtype?
Discrepancies between the IHC and gene panels have been studied in numerous recent studies: Paquet and Hallett found a 28.6% discordance between the PAM50-based intrinsic subtype and the IHC immunophenotype.6
Hee Kyung Kim found discordance in 38.4% of cases when comparing PAM50 with the molecular subtype surrogated by IHC. Only 29% of HER2-enriched patients (PAM50) were HER2+ by IHC. The authors commented that such discordance may be a cause of under- or over-treatment since HER2-enriched/HER2-negative IHC patients (71%) could confer lack of potential benefit to monoclonal therapy.7 Although the St. Gallen Conference 2011 recommended treating patients based on the IHC assessment of ER, PR, HER2 and Ki-67, the results of this study suggest that these discrepancies should be taken into account.
In the study by Parker et al.,8 the authors compared the molecular subtype by PAM50 with the IHC surrogate and observed that, of 626 RE+tumours (IHC), 73% were Luminal (A or B), 11% were HER2-enriched, 5% were Basal-like and 12% were Normal-like. In contrast, RE- (IHC) tumours included 11% Luminal, 32% HER2-enriched, 50% Basal-like and 7% Normal-like. That is, the authors observed discordances of 45.1% in the Luminal group, with Luminal A accounting for only 58.3% of the HR+/HER2- tumours. Of the HR−/HER2+tumours, 80% were HER2-enriched and of the HER2-positive (IHC) tumours, only 51% were HER2-enriched by PAM50.
Viale et al., in a comparative study between IHC and BluePrint®,9 observed that BluePrint® reclassified 30% of the cases. The group with the greatest discordance was the Luminal tumours and the most concordant was the Triple-negative tumours (TN). BluePrint® reclassified 54% of the Luminal B (IHC) tumours as Luminal A tumours (Low Risk), 38% of HER2+tumours (IHC) as Luminal (A or B) tumours and 5% as Basal tumours. Of the Triple-negative IHC group, 5% were reclassified by BluePrint® as Luminal (A or B).
Taking these data together, we observe that the greatest discrepancies are in the Luminal group (between low and high risk, Luminal A vs. Luminal B) and in the HER2-enriched group.
These results suggest, on the whole, that patients with the same IHC surrogate subtype have important genetic differences and that their immunophenotypic classification may result in inadequate treatment and poor survival, which raises the need to review current guidelines to ensure the correct intrinsic characterisation of tumours.
To explain the discrepancies, different arguments have been put forward and many studies attribute these discrepancies to alterations in tissue fixation, antibody choice and variations in interpretation. Hammond et al. noted that up to 20% of oestrogen and progesterone receptor results may be inadequate10 while other studies report that the rate may be even higher.11,12
Other studies, however, found high rates of concordance between IHC and microarray gene expression, considering their use exclusively for the molecular classification of breast carcinoma.13–17
Another potential cause of discordance between GS and IHC is attributable to the Ki-67 proliferation factor. One of the criteria for discriminating between low and high risk is the Ki-67 factor, whose reproducibility is high for <30 and >60 percentages, but poor for the intermediate range. In addition, the cut-off points for Ki-67 have changed over time. In 2011, the St. Gallen Expert Group proposed a cut-off point of 14%18 but, two years later, Cheang et al.19 proposed to change this to 20%, given the poor reproducibility of the initial cut-off point.
With respect to the IHC reproducibility of HER2, although both the IHC test and the FISH test have shown good specificity and good sensitivity in most cases, some issues are yet to be resolved, such as the correct classification of equivocal cases (approximately 12%), identification of “HER2-low” cases, and early prediction of resistance to monoclonal therapy.20
Other authors21 compared the “clinical subtype” (IHC/FISH) with MammaPrint®/BluePrint® subtyping in a series of 474 patients to predict treatment sensitivity, reclassifying 87 of the 474 (18%) patients as Basal-type with a pathological complete response rate of 32% versus 11% of “clinical” Luminal tumours.
- 2.
The second question, regarding risk prediction for individual cases, is whether the risk of recurrence and metastasis (and, consequently, the decision on whether or not to prescribe chemotherapy) assigned by different GSs is the same or may vary depending on which one we apply. In other words, are the different genomic platforms classified in the same risk category?
The study by Bartlett et al.22 showed a 60% discrepancy in the level of risk between the different platforms compared (MammaPrint®, Oncotype DX®, PAM50, IHC4, IHC4-AQUA). These differences are attributable to the difference in the genes included in each platform, the different technologies involved and the different mathematical methods applied in their development, but it is important to know whether these differences impact on the prognosis of individual patients. In the study by Cheng,23 it was observed that, at a general level, despite the poor concordance between the genes included in the different platforms evaluated, the different models offer similar predictions since they reflect common cellular phenotypes that encompass the differences between the ER+luminal cancer and the ER- (basal-like, HER2+and ER-negative) cancer.
At an individual level, comparative data from the different platforms, limited and based on the in silico reconstruction of existing evidence, suggest only a “moderate” degree of concordance, with the discrepancies between individual predictions being statistically significant. The question is, however, to some extent rhetorical given that not all GSs include the same risk categories (MammaPrint®, EndoPredict®: High and Low Risk, Oncotype Dx® and PAM50: High, Intermediate and Low risk).
Recently, the PROMIS study, whose objective was to re-evaluate, using MammaPrint®, a series of 840 patients classified as intermediate risk by the 21-gene platform, reclassified 44.5% as low risk and 55.5% as high risk. This resulted in the modification of the therapeutic decision in 33.6% of the patients included in the study after receiving the results from the 70-gene platform, avoiding chemotherapy in 28.9% of low-risk MammaPrint® patients and adding chemotherapy for 36.7% of patients classified as high risk by the 70-gene platform. In conclusion, the study shows that there are indeed differences in the assigned risk group depending on the genomic platform applied. Orucevic's study24 on the impact of the intermediate risk category, whose PPV was only 40.1%, reflects the lack of therapeutic recommendations for this patient group.
What is the reason for these differences? In our view, one of the key reasons for this is the small or no overlap between the genes included in the different platforms (between MammaPrint® and Oncotype Dx® there is only a single gene match), which can lead to variation in the assigned risk group. In conclusion, we believe that the use of genomic platforms improves therapeutic decision-making in almost one third of cases.
The PROMIS study reports increased physician confidence in MammaPrint® following the reclassification of intermediate-risk cases by Oncotype Dx®, and observed an 88% adherence to recommendation for the test in RH+patients requiring chemotherapy and 91% in low-risk patients likely to avoid chemotherapy.
Moreover, the IMPACT prospective study25 compared the strategy recommended by the physician before and after receiving the results of the MammaPrint®/BluePrint® test for a series of 358 patients, observing modification of the therapeutic decision in 24% of the cases. The study also noted that, in 72% of the cases included, the patients had greater confidence in the proposed treatment plan after receiving the results of the GS.
Question 8. Do you think there is an advantage to performing the test at the hospital itself?More than half of those surveyed (53.8%, 14 answers) considered that having the platform in the hospital itself offered a quick response and optimisation of resources. 8% of the respondents (2 answers) believed that it improved the understanding of the result and 27% (7 answers) did not see any advantage to it.
In our opinion, more important than the origin of the information provided by the GS, whether internal or external, is the quality of that information and its inclusion in the pathology report so that it can be recorded in the patient's medical history. In addition, the results of the GS should be evaluated by the pathologist, compared with the immunohistochemical immunophenotype of the tumour, not always coincident, and contextualised with the morphological findings and the patient's medical data. This information should be discussed in the tumour committee in order to make therapeutic decisions based on the best possible knowledge, with the current tools, of tumour biology.
Question 9: Do you think the GS has any impact on axillary treatment? 62% of respondents (16 answers) said ‘no’, compared to 38% (10 answers) who said ‘yes’.With regard to the answers, it would be interesting, in our opinion, to know how the question was interpreted. Our comments are structured along five different axes:
- 1.
Axillary staging and molecular subtypes: Currently, we rarely indicate axillary treatment based on molecular type, except for elderly patients with early-stage, receptor-positive tumours for whom lymphadenectomy, and even the sentinel lymph node, are known to have no survival benefit.
In a prospective, randomised study involving 473 patients of ≥60 years with T1–T3 tumours, with a mean age of 74 years, the patients were assigned to surgery with lymphadenectomy versus surgery without axillary clearance followed by tamoxifen and a 5-year follow-up. The primary objective was quality of life. With a mean follow-up of 6.6 years there was no difference between disease-free survival (clearance group 67%, non-clearance 66% CI 0.79–1.42, p=0.69) and overall survival (clearance 75% non-clearance 73% CI 95% 0.76–1.46 p=0.77). With regard to the quality of life questionnaires, these were more positive for the group without axillary clearance.26
Similar results were seen in another randomised study involving 219 T1 N0 node-negative patients aged 65–80 years, with positive hormone receptors (88%) and treatment with tamoxifen. No statistically significant differences were found in mortality, events in the breast or axilla, or overall survival. These results led the authors to conclude that axillary dissection can be omitted in patients with early-stage breast cancer without palpable nodes. They further concluded that the sentinel lymph node would also not provide any benefit to elderly patients with positive receptors.27
Therefore, leaving aside this special group of elderly patients with positive hormone receptors in whom we could consider that molecular subtypes can influence axillary surgery, in the rest of the cases we currently have no evidence for deciding on axillary surgery based on molecular subtype.
- 2.
Axillary involvement and molecular subtypes: Another aspect is to consider whether the molecular type affects the rate of lymph node involvement. There is evidence that the classification categorised into the 5 subtypes is indeed related to the rate of axillary involvement. In a study of 2984 tumours classified into molecular subtypes, following adjustment for confounding factors, it was possible to show that the intrinsic subtype was a predictor of lymph node involvement in luminal B HER2 (OR=1.49, p=0.09), overexpressed HER2 (OR 1.61, p=0.015) and Basal-like (OR 0.60, p=0.02)28 tumours. It is also known that despite the poor prognosis and higher probability of local recurrence of triple-negative tumours, they have a lower probability of lymphovascular invasion and lymph node involvement, which suggests that these types of tumours metastasise less frequently via the lymphatic route. This was observed in a series of 11,596 tumours that were classified into surrogate types. Adjusting for the other variables, it was evident that luminal and HER2 tumours had more vascular invasion and a greater probability of having >4 lymph nodes than triple-negative tumours.29
Thus, recognising that nodal involvement and molecular classification are two important prognostic factors, it is relevant to study the direct relationship between the two aspects and the prognostic value of nodal involvement in the different molecular subtypes.
In a study with 4262 patients, it was shown that luminal A-like tumours and triple-negative tumours are less likely to have nodal involvement and a pN3 nodal stage than luminal B and HER2 tumours.30 Although some other studies seem to show no relationship between molecular subtypes and nodal involvement,31 it seems that nodal status and molecular subtypes play an important role as prognostic factors in patients with breast cancer.
- 3.
Choice of primary treatment: On completion of the diagnosis and extension study, two aspects are considered when choosing the primary treatment: the TN and molecular subtype. In general terms, from a certain size (2cm or even less) and with nodal involvement, the indication for primary treatment with either chemotherapy or hormone therapy is assessed with the aim of testing the chemo or hormone sensitivity of the tumour, reducing the staging to perform a less aggressive surgery and attempting a complete pathological response that could become an independent prognostic factor. In this way, the molecular subtype becomes a predictive factor for treatment.
Thus, if we take the molecular subtype as a factor when deciding on a neoadjuvant treatment, it is important to consider that there are discrepancies between the surrogate molecular subtypes and the profiles from genomic platforms.
Surrogate types and gene expression profiles: In a study of 426 patients aimed at comparing the molecular subtype by immunohistochemistry and fluorescence in situ hybridisation, versus Mammaprint®/BluePrint® genomic platforms as a predictor of chemosensitivity, a different reassignment was observed in 22% of the cases. 42% of HER2 tumours by immunohistochemistry were reclassified into other groups, 98% of triple-negative tumours were confirmed as basal tumours by BluePrint®, and 96% of luminal tumours were confirmed by BluePrint®. Following reclassification, the two groups in the study that improved in terms of complete response rates were HER2 tumours and the luminal tumours that were reclassified as basal tumours.32 We should therefore ask ourselves whether, given these discrepancies, we would need to broaden the indications of the genomic platforms in order to make a more targeted choice of primary treatment.
- 4.
Genomic platform for predicting the response to neoadjuvant treatment. Until now, in the care setting, we have made restrictive use of genomic platforms. Thus, their use is most often restricted to those cases where the benefit of adjuvant chemotherapy is doubtful, either because of tumour size, nodal involvement, or borderline prognostic or discordant factors. The platform's assignment of high recurrence risk helps us to decide on the chemotherapy treatment despite the possible associated side effects. An expanded use would be to use genomic platforms as a predictor of response to neoadjuvant treatment.
- ∘
Neoadjuvant chemotherapy: Tumours with positive hormone receptors have a highly heterogeneous biology and represent a challenge for accurate treatment choice. In the Neoadjuvant Breast Registry Symphony Trial, 474 tumours were classified with Mammaprint®/Blueprint® and compared with the previous classification by immunohistochemistry and the complete response rates to primary chemo or hormone therapy. MammaPrint® classification had a higher probability of a complete response than immunohistochemistry, and 18% (87/474) of luminal tumours were reclassified into different subgroups (1 HER2, 86 basal). Patients with basal tumours by BluePrint® had complete response rates of 32% to chemotherapy, while BluePrint® luminal tumours had a partial clinical response rate of 68% to hormone therapy.32,33
- ∘
Neoadjuvant hormone therapy: The predictive use of genomic platforms has also been applied in the primary hormone therapy setting. This is how the biopsies of 87 patients with hormone-positive tumours who had received hormone therapy such as tamoxifen or anastrozole were studied. The 21-gene expression profile (Oncotype Dx®) was applied in order to study whether the recurrence score (RS) predicted the response to primary hormone therapy. Despite the lack of material in the biopsy, in half of the cases it was evident that if the RS was low there was a greater response rate to primary hormone therapy than if it were intermediate or high.34 In the same vein, the TranNeos study evaluated whether the Oncotype Dx® RS was able to predict the response to primary hormone therapy. The primary objective was to objectify the clinical response (complete or partial) to neoadjuvant letrozole for RS<18 (55%) versus 22% for RS≥31. In addition, it was observed that patients with a RS>18 were more likely after neoadjuvant treatment to have conservative surgery versus patients with RS≥31.35
- ∘
- 5.
Axillary surgery and genomic platform: The evolution of axillary surgery, the development and consolidation of the sentinel lymph node technique, and paradigm changes such as that brought about by the ACOSG Z0011 study36 have completely changed the current concept of axillary surgery. Axillary surgery is a method of staging, preserving tumour debulking in axillae with a high axillary tumour burden and in chemo or hormone-resistant disease. Thus, if the genomic platforms are predictive of response to chemotherapy or hormone therapy, in cases of axillary involvement, we must achieve a complete axillary response to reduce the aggressiveness of surgery and avoid axillary leakage.
19.2% of the respondents (5 answers) believe that it can at 5 years while 76.9% (20 answers) responded that the GS can evaluate the risk of a relapse at 10 years. 3.8% (1 answer) answered that the GS can evaluate the risk of relapse after more than 10 years.
Knowing the biology of breast cancer is the key to adjusting treatments to the risk of recurrence and avoiding overtreatment. For some decades we have been basing ourselves on clinical and histological parameters that have allowed us to define the surrogate types to indicate the different therapies. Gene expression platforms represent a further step towards personalising treatments and defining high-risk patients who will need precise treatments despite their aggressiveness, as opposed to those with indolent, slow-growing tumours with a very low probability of recurrence, to whom we will need to apply therapies with fewer side effects, or simply establish close, structured monitoring. And this aspect is even more important, since population screening has increased the diagnosis of low-risk, indolent tumours, to which we currently apply the same treatments as for other high-risk tumours.37
The biological heterogeneity of breast cancer is so varied that the latency time to recurrence can range from a few months to several decades after diagnosis and treatment. Genomic platforms identify tumours with a risk of early recurrence and patients who would benefit from chemotherapy treatment to reduce this risk.38 A secondary analysis of the STO-3 study included 652 menopausal patients divided into two groups according to whether they had been treated with tamoxifen (339) or not (313). These patients were classified by MammaPrint® as low risk (58%) and high risk (42%). With the MammaPrint® platform, 98 (15%) ultralow risk patients were identified. With a 20-year follow-up, patients classified as low or high risk, but not ultralow risk, had a higher and statistically significant percentage of cancer-specific mortality. The 15-year specific survival rates of ultralow risk patients in the tamoxifen-treated group was 97%, and 94% in the untreated group. No deaths were reported in the tamoxifen-treated ultralow risk group. With regard to the characteristics of the ultralow risk tumours, all were hormone receptor-positive HER2-negative, and luminal tumours by BluePrint. 96% had a Ki-67 of<15% and 89% were luminal tumours by PAM50. In contrast, only 19% of hormone receptor-positive HER2-negative tumours and 20% of low Ki-67 tumours were ultralow risk. In addition, 25% of the luminal tumours by PAM50 and 26% by BluePrint were ultralow risk.39 The patients in this group had such a good prognosis that, if we were able to identify the precursors of these tumours, when they were diagnosed by screening we could have considered therapeutic abstention and monitoring. Thus, we can conclude that immunohistochemistry is unable to select these indolent growth, low-risk tumours, and that only the genomic platform can identify them, with the therapeutic advantages that can be derived from avoiding overtreatment and its associated morbidity.
PerspectivesGene expression profiles allow the identification of molecular subtypes in breast cancer. However, defining cancer biology on the basis of gene expression alone and without knowledge of related changes in the proteome remains a challenge, since proteins are the functional key to biology as well as therapeutic targets of anti-cancer drugs.
Recent technological advances in mass spectrometry have revealed ideas that had not been revealed by mRNA-based studies, giving “proteo-transcriptomic” analyses the potential to uncover previously undescribed molecular alterations in cancer biology.
A recent study40 reveals that proteome and transcriptome describe different areas of tumour biology and that the proteins are more frequently over-regulated than their corresponding transcripts. Thus, mRNA abundance incompletely predicts protein abundance in breast cancer tissue, and even less so in adjacent non-cancerous tissue. Furthermore, the disease patterns described by the proteome were only partially captured by the tumour transcriptome, coinciding with the results from the NIH CPTAC group.
The study discovered that protein-mRNA concordance in breast cancer is a characteristic, new prognostic factor associated with molecular subtype, aggressiveness and lower survival. Few studies investigate whether the proteomic signature of cancer allows for classification into prognostic subgroups.
This and other lines of research are still necessary to advance in the understanding, diagnosis and treatment of a complex disease such as breast carcinoma.
ConclusionsImplementation of genomic platforms impacts on decision-making by modifying the therapeutic recommendation and generally reducing the number of patients treated with adjuvant chemotherapy. Our survey was exploratory and therefore its value is only indicative. Although it does not allow definitive conclusions to be drawn, given the small size of the sample, it did serve as a starting point for clarifying some conclusions based on scientific evidence, useful, in our opinion, for consolidating basic concepts.
- •
The indications for GS, financed by the Public Administration, must be rethought and expanded, in the light of new evidence, in positive lymph nodes and, in the very near future, in the neoadjuvant setting.
- •
Although all respondents said they knew about and believed in the evidence for decision-making, we often do not know the facts of this evidence and are not consistent in using it.
- •
MammaPrint® is the only platform with an evidence level of 1 for use with N1 patients.
- •
Immunohistochemistry and genomic signature do not provide interchangeable information, but rather complementary information.
- •
The use of genomic signatures increases the confidence of both the physician and the patient in the therapeutic decision.
- •
Currently, there is not enough evidence for the molecular subtype to be decisive in making decisions regarding the axilla.
- •
MammaPrint® identifies a subgroup of patients with “ultralow risk” breast cancer, not identifiable using other diagnostic methods such as immunohistochemistry or FISH, who may likely not receive any treatment.
- •
Although genomic platforms are already a commonly used tool in the classification and treatment of breast cancer, the evolution of knowledge will make it possible to offer innovative treatments based on a better understanding of tumour biology.
The authors have not received any funding for this study.
Confidentiality of dataThe characteristics of the study exempt it from being evaluated by a bioethics committee.
Conflict of interestLaia Bernet is associated editor of Revista de Senología y Patología Mamaria. The rest of the authore declare not to have conflict of interest.