Abuse and misuse of antimicrobial agents has accelerated the spread of antimicrobial-resistant bacteria. The association between antimicrobial-resistant infections in humans and antimicrobial use in agriculture is complex, but well-documented. This study provides a systematic review and meta-analysis of the dissemination of antimicrobial resistance (AMR) to antimicrobials defined as critically important by the WHO, in swine, chicken, and cattle from intensive and extensive production systems in Argentina. We conducted searches in electronic databases (MEDLINE-PubMed, Web of Science, SciELO, the National System of Digital Repositories from Argentina) as well as in the gray literature. Inclusion criteria were epidemiological studies on AMR in the main food-transmitted bacteria, Salmonella spp., Campylobacter spp., Escherichia coli and Enterococcus spp., and mastitis-causing bacteria, isolated from swine, chicken, dairy and beef cattle from Argentina. This study gives evidence for supporting the hypothesis that AMR of common food-transmitted bacteria in Argentina is reaching alarming levels. Meta-analyses followed by subgroup analyses confirmed the association between the prevalence of AMR and (a) animal species (p<0.01) for streptomycin, ampicillin and tetracycline or (b) the animal production system (p<0.05) for streptomycin, cefotaxime, nalidixic acid, ampicillin and tetracycline. Moreover, swine (0.47 [0.29; 0.66]) and intensive production (0.62 [0.34; 0.83]) showed the highest pooled prevalence of multidrug resistance while dairy (0.056 [0.003; 0.524]) and extensive production (0.107 [0.043; 0.240]) showed the lowest. A research gap regarding beef-cattle from feedlot was identified. Finally, there is an urgent need for political measures meant to coordinate and harmonize AMR surveillance and regulate antimicrobial use in animal production.
El abuso y mal uso de los antimicrobianos aceleró la propagación de bacterias resistentes. La asociación entre las infecciones que presentan resistencia a antimicrobianos (RAM) en humanos y el uso de antimicrobianos en la producción agropecuaria es compleja, pero está bien documentada. Proporcionamos una revisión sistemática y metaanálisis sobre la diseminación de la resistencia a antimicrobianos designados como críticamente importantes por la Organización Mundial de la Salud (OMS) en cerdos, aves y bovinos de producción intensiva y extensiva en Argentina. Se buscó información en bases de datos electrónicas (Medline-PubMed, Web of Science, SciELO, Sistema Nacional de Repositorios Digitales de Argentina) y en la literatura gris. Se incluyeron estudios epidemiológicos sobre la RAM en las principales bacterias transmitidas por los alimentos – Salmonella spp., Campylobacter spp., Escherichia coli y Enterococcus spp. – y bacterias causantes de mastitis aisladas de cerdos, pollos y bovinos. Los resultados de este estudio apoyan la hipótesis de que la RAM de las bacterias transmitidas por los alimentos alcanza niveles alarmantes. Los metaanálisis seguidos de análisis por subgrupos mostraron asociación entre la RAM y (a) el animal (p<0,01) para estreptomicina, ampicilina y tetraciclina o (b) el sistema productivo (p<0,05) para estreptomicina, cefotaxima, ampicilina, ácido nalidíxico y tetraciclina. La mayor prevalencia conjunta de multirresistencia se detectó en cerdos (0,47 [0,29; 0,66]) y producción intensiva (0,62 [0,34; 0,83]), mientras que la menor correspondió a bovinos de leche (0,056 [0,003; 0,524]) y producción extensiva (0,107 [0,043; 0,240]). Se observó un vacío de información respecto de los bovinos de feedlot. Es urgente adoptar medidas políticas para coordinar y armonizar la vigilancia de la RAM y regular el uso de antimicrobianos en animales.
The discovery of antimicrobials has been one of the main advances in the history of medicine30. The appropriate use of antimicrobials is important in ensuring human and animal health as well as welfare, food safety and food security27,64. However, abuse and misuse of antimicrobials has allowed the global dissemination of antimicrobial resistance (AMR) in clinical and non-clinical species61,96.
A well-known case of inappropriate use of antimicrobials is represented by livestock farming where much of its global use is to fulfill non-therapeutic purposes such as growth promotion and infection prevention2,7,9. The world population growth coupled with a higher purchasing power in low- and middle-income countries has driven an increase in the demand for high-value animal protein88. Therefore, the main food-producing countries, including Argentina, have opted for highly profitable and vertically integrated intensive production systems, which rely on the use of antimicrobials to maintain productivity and animal health78.
There is compelling scientific evidence available to support that antimicrobial use on farms imposes a public health burden due to infections with resistant pathogens37. Moreover, transference of AMR genes to the human gut microbiome can also occur through the food chain17,38,69. Despite these risks, the use of antimicrobials in animals has remained largely unregulated49.
If no political measures are taken to stop the spread of AMR, it is expected to cause 10 million deaths per year toward 2050, more than cancer-related deaths60,63. Given the dry pipeline for new antimicrobials, we must expertly manage the drugs that are currently available30. Therefore, the World Health Organization (WHO) in collaboration with the Food and Agriculture Organization (FAO) and the World Organization for Animal Health (OIE) have declared AMR an issue of global concern which does not recognize geographical nor human/animal boundaries and therefore, requires a holistic and multi-sectoral approach95.
In June 2015, the Argentine Ministries of Health and Agriculture, Livestock and Fisheries approved the Argentine Strategy for Control of Antimicrobial Resistance, and the National Commission for the Control of Antimicrobial Resistance was created (CONACRA, https://www.argentina.gob.ar/tags/conacra). As part of the action plan, an AMR surveillance program has begun. It started to analyze the prevalence of AMR of importance for public health, among commensal and pathogenic bacteria isolated from fecal samples obtained in slaughterhouses. In addition, the national registrations and certificates for the use and commercialization of animal feed containing antibiotics, antiparasitics or coccidiostats were automatically deregistered as of January 2019 (SENASA Resolution No. 1119/18). Moreover, since that year, the production, distribution, imports, use and possession of veterinary products containing colistin were also banned in Argentina (RESOL-2019-22-APN-PRES#SENASA). However, the use of certain antimicrobials as growth promoters for animal production is still allowed in this country (Luna F., personal communication, 2022, SENASA).
To date, there are no comprehensive studies that systematically document the dissemination of AMR in the rapidly growing, intensive animal production industry being developed in Argentina33, a country with a longstanding tradition of extensive animal production systems (MAGYP, https://www.magyp.gob.ar/sitio/areas). Given the leading role of livestock industries in the economy of Argentina, representing approximately USD 4000 million exported to different markets yearly40, the revision of AMR mechanisms in this area is mandatory.
Therefore, our objective was to document the dissemination of AMR to antimicrobials ranked as critically important for human medicine by the WHO, among commensal and pathogenic bacteria isolated from food-producing animals from Argentina, contrasting intensive and extensive production systems. These findings may help to develop highly relevant policies and regulations not just for Argentina but for other countries undergoing similar intensive animal production growth.
MethodologySearch strategyA systematic search was conducted in the international databases MEDLINE-PubMed (ncbi.nlm.nih.gov/pubmed/), Web of Science (webofknowledge.com), SciELO (scielo.org/en/), and the National System of Digital Repositories of Argentina (repositoriosdigitales.mincyt.gob.ar/vufind/). The search strategy included the terms “cattle”, “dairy”, “swine”, “pig”, “chicken”, “anti-infective agent resistance”, “antimicrobial resistance”, and “Argentina”. The last search was performed on January 24th 2022.
In addition, generic and academic searches were conducted in the Internet in order to identify relevant gray literature including conference papers and government documents. When necessary, the authors were contacted to obtain further information about their studies. Key informants from the National Service of Agri-Food Health and Quality (SENASA) were also contacted.
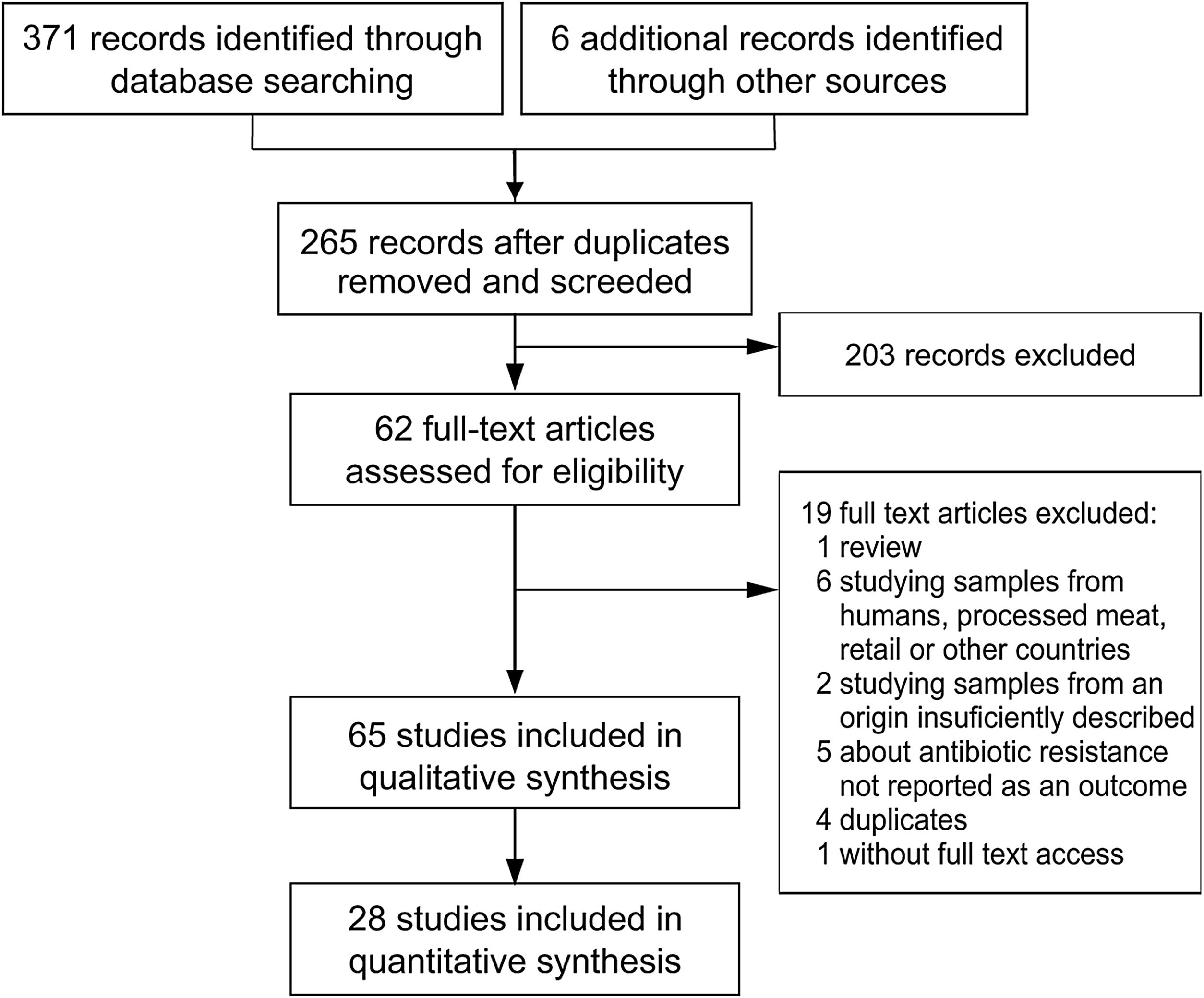
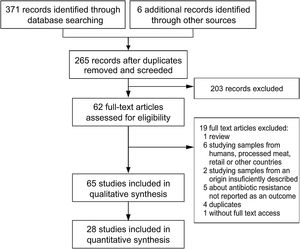
Study selectionStudy selection was performed following the flowchart reported by Moher et al.53 (Fig. 1). The first screening was performed at a title and abstract level, and the full texts of the records retained were assessed for eligibility. Those included were epidemiological studies about AMR in Salmonella spp., Campylobacter spp., Escherichia coli and Enterococcus spp., the main four food-transmitted bacteria isolated from swine, chicken, dairy and beef cattle, whose products were intended for human consumption, published in English or Spanish and which included the assessment of animals coming from farms in Argentina. Given that clinical mastitis was reported to be the most prevalent disease in adult dairy cows, which together with dry cow therapy, represent 85.4% of total drug usage at herd level34, when the report was on dairy, mastitis-causing bacteria were also included. To be as inclusive and comprehensive as possible, there were no limits placed on study quality, with study quality issues handled through sensitivity analyses instead.
Finally, for the meta-analyses, studies that presented recovery strategies with antimicrobial selection or, selection based on the presence of an AMR genetic determinant were not considered eligible. Moreover, exclusively for the meta-analyses of food-transmitted bacteria, studies involving targeted sampling of animals with clinical symptoms were not considered eligible either.
Data extraction and definitionsWhen different animal species or bacterial genera had been included in a scientific article, each animal species or bacterial genus was included separately in the review as a particular “study”. Similarly, when a scientific article reported the results derived from different animal production systems, each one was considered as an individual study. Therefore, a single scientific article could represent several included studies. By contrast, when a scientific article reported results from different stages of animal breeding in the same farm, it was considered as a single study.
Data from the full text of the selected primary studies were extracted by using Excel spreadsheet, and graphics were made using Excel, Power Point, and the statistical software R-4.1.2 (2021-11-01). Information about geographical distribution, year of publication, food-producing animals sampled, animal production system (intensive, confined, extensive, family farming), type of samples, number of samples, sampling period, bacteria assessed, methodology to determine AMR, and the outcomes (number of isolates resistant to critically important antimicrobials and total isolates analyzed) were extracted from each article.
The definitions followed throughout the article were, prevalence: number of isolates over total samples analyzed; prevalence of resistance to critically important antimicrobials: number of isolates resistant over total isolates analyzed; and multidrug resistance (MDR): number of isolates resistant to three or more antimicrobial classes over total isolates analyzed48. Isolates with intermediate susceptibility were classified as susceptible. When the definitions followed by the studies differed from the ones presented here, if possible, the outcomes were re-calculated.
For production system comparison, systems were classified as ‘intensive’, ‘extensive’ or ‘not described’. A study was assigned to the category ‘intensive’ if animal husbandry was classified by authors as intensive, industrialized and/or where animals spend most of their life confined indoors. On the contrary, a study was assigned to the category ‘extensive’ if animal husbandry was classified by the authors as extensive, free-range, backyard, family farming and/or where animals spend most of their life grazing outdoors. Studies with not enough information to classify their systems as ‘intensive’ or ‘extensive’ were considered ‘not described’.
To simplify the AMR report we adopted the following terms to describe the levels of AMR occurrence: ‘rare: <0.1%’, ‘very low: 0.1% to 1.0%’, ‘low: >1% to 10.0%’, ‘moderate: >10.0% to 20.0%’, ‘high: >20.0% to 50.0%’, ‘very high: >50.0% to 70.0%’, ‘extremely high: >70.0%’.
Statistical analysisA statistical analysis was performed using the statistical software R-4.1.2 (2021-11-01) with the {meta}6 and {dmetar}50 packages. Meta-analyses were conducted separately for each group of bacteria and each antimicrobial. Particularly for food transmitted bacteria, meta-analyses were followed by subgroup analyses to explore the sources of heterogeneity. Each meta-analysis was only performed if there were six or more studies, and each subgroup analysis was only done if there were 10 or more studies, to reduce the risk of obtaining biased pooled estimates35.
The binary parameter ‘proportion’ (with 95% confidence intervals) was the one selected to measure effect size35. We pooled logarithmic proportion of resistant isolates by antimicrobial, using the Hartung–Knapp correction for random-effects model with the function metaprop. The method specified was inverse-variance pooling. The function forest.meta was used to construct Forest Plots.
Between-study heterogeneity was assessed using Cochran's Q test and the inconsistency index (I2-statistic). Sensitivity analyses were performed with the function InfluenceAnalysis from {dmetar}50 to test for highly influential studies. The function performs the same analysis, but leaving-one-out in each iteration. Subgroup analyses were performed using the random-effects model, assuming common tau^2 in the subgroups35. For food-transmitted bacteria, a priori subgroup analyses were planned to assess the association between AMR and (1) food-producing animals or (2) food-producing systems. A posteriori, a subgroup analysis for (3) bacteria was included, which was necessary because, based on the reduced number of studies, results from Salmonella spp. and E. coli were pulled together under the category Enterobacteriaceae.
The presence of publication bias was assessed using funnel plots with the function funnel.meta from {meta}6. Funnel plot asymmetry was tested using the Egger's regression test with the function metabias23. This test was only performed in meta-analyses with 10 or more studies.
ResultsExcluded studiesThe literature search yielded two hundred and sixty-five records after duplicates were removed (Fig. 1). Reviews, studies on parasites or viruses, studies about samples from humans, processed meat, retail or other countries, studies on the development of new drugs or new treatments, studies about alternative growth promoters or methods and studies where AMR was not tested or tested only by PCR, were excluded.
Overview of included studiesForty-three articles of two hundred and sixty-five screened were eligible for qualitative synthesis (Fig. 1). Among these articles, the first one was published in 1997, two were subsequently published until 2000, nine between 2001 and 2010, twenty-seven between 2011 and 2020, and the remaining four in 2021 (Fig. 1).
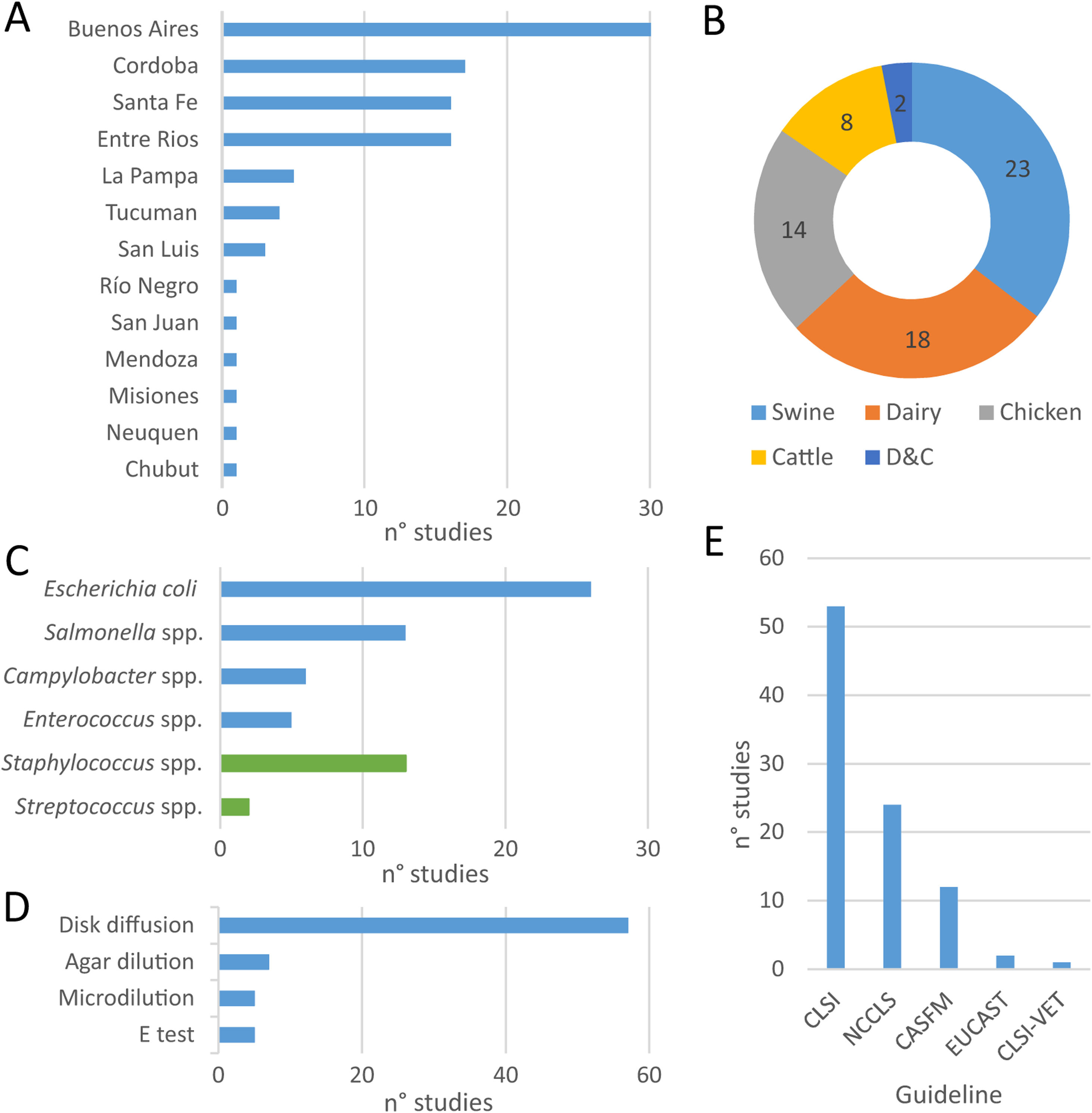
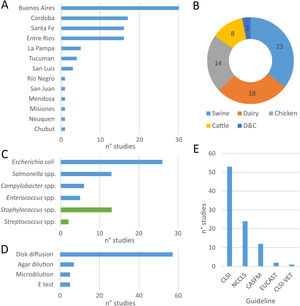
Within the eligible articles a total of sixty-five studies was identified, differing in geographical distribution, subjects studied, animal production systems and, methods and guidelines followed to identify AMR (Fig. 1). Of Argentina's twenty-three provinces, the data came mainly from those in the center region of the country. Buenos Aires province was the most represented one, followed by the provinces of Cordoba, Santa Fe and Entre Ríos (Fig. 1A).
At least one study was found about each food-producing animal (chicken, swine, beef-cattle and dairy), food-transmitted bacteria (E. coli, Enterococcus spp., Salmonella spp., Campylobacter spp.) and the main mastitis-causing bacteria (Fig. 2B and C). The largest proportion of studies assessed swine3,8,12,13,24,39,42,45,56–59,65,66,89,91–93, followed by dairy11,12,16,31,32,36,44,52,55,58,67,68,71,73,79–83,89, chicken14,18–20,62,65,72,86,98 and beef cattle12,44,52,65,79,89 (Fig. 2B). Among mastitis-causing bacteria, all isolates belonged to genera Staphylococcus11,31,32,55,68,71,73,80–83 and Streptococcus16,36. While E. coli was the most frequently studied food-transmitted bacteria3,8,12,18,19,24,44,45,52,56–59,65,67,79,86,89 (Fig. 2C), Staphylococcus was the most studied mastitis-causing genus (Fig. 2C).
Geographic distribution of studies on antimicrobial resistance included in this review (A), coverage of food-producing animals (B) and bacteria (C), methods used (D) and guidelines followed (E) for antimicrobial resistance testing and interpretation of results. More information about each of the studies is available in Suppl. Table S1.
With respect to the methods applied to test susceptibility to antimicrobials, disk diffusion was the one preferred by almost all authors (Fig. 2D). The main exception was broth microdilution3,18,19,55,71, which was particularly needed to test susceptibility to colistin. Conversely, the E-test was used as a complement, but never as an only option16,62,73,82. Similarly, when comparing guidelines, all authors followed NCCLS guidelines – before 2008 – or CLSI guidelines – after 2008 – except for particular antimicrobials (Fig. 2E). For example, CASFM guidelines were followed by some authors when assessing susceptibility to streptomycin65 and EUCAST guidelines when assessing susceptibility to colistin18,19.
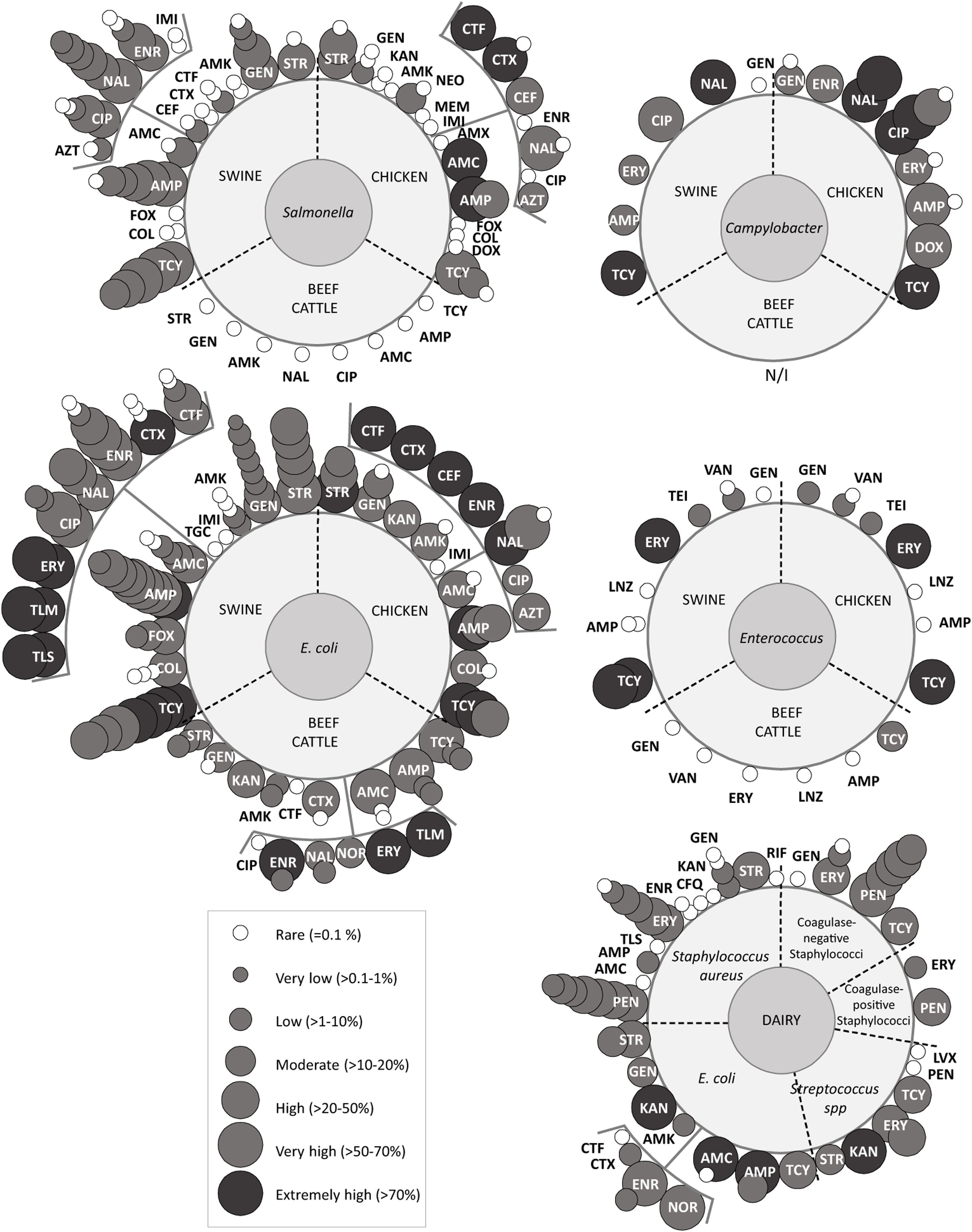
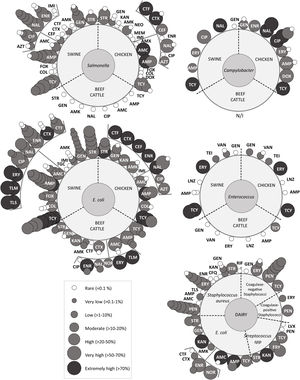
Overview of antimicrobial resistanceReports on AMR were initially divided into five groups which included the following bacteria (Fig. 3). Group ‘Campylobacter’ included Campylobacter spp.65,98 and Campylobacterjejuni62; group ‘Enterococcus’ only included Enterococcus spp.14,65,91; group ‘E. coli’ included E.coli3,18,24,56,58,59,65,79,86,89, Shiga toxin-producing E.coli12,44,52, enterotoxigenic E.coli57, colistin-resistant E.coli19 and integron positive E.coli12,45; group ‘Salmonella’ included Salmonella spp.13,65,72,86,92,93, Salmonellaenterica42,66, Salmonella enterica subspecies enterica39 and Salmonella Heidelberg13,20; and group ‘dairy’ included E.coli79,89, Shiga toxin-producing E.coli12,44, enterotoxigenic E.coli67, integron positive E.coli12, and mastitis-causing bacteria Staphylococcusaureus31,55,68,73,80,81,83, coagulase-negative staphylococci32,71,80,82,83, coagulase-positive staphylococci11, Streptococcus spp.16 and Streptococcusagalacteae36.
Antimicrobial resistance rates to critically important antimicrobials reported for each bacteria and food-producing animal included; overlapped spheres represent AMR reported by different studies. AMK: amikacin; AMP: ampicillin; AMX: amoxicillin; AMC: amoxicillin+clavulanic acid; AZT: aztreonam; CEF: cefepime; CFQ: cefquinome; CIP: ciprofloxacin; COL: colistin; CTF: ceftiofur; CTX: cefotaxime; DOX: doxycycline; ENR: enrofloxacin; ERY: erythromycin; FOX: fosfomycin; GEN: gentamicin; IMI: imipemem; KAN: kanamycin; LVX: levofloxacin; LZD: linezolid; MEM: meropenem; NAL: nalidixic acid; NEO: neomycin; NOR: norfloxacin; PEN: penicillin; POL: polymyxin; RIF: rifampin; STR: streptomycin; TEI: teicoplanin; TGC: tigecycline; TLM: tilmicosin; TLS: tylosin; TCY: tetracycline; VAN: vancomycin. This figure includes data from articles that presented recovery strategies with antimicrobial selection or, selection by the presence of a genetic determinant of AMR, or targeted sampling of animals with clinical symptoms. Studies about E. coli with less than 4 isolates were not represented in the figure. More information is available in supplementary data.
The overview of antimicrobials tested included almost every class classified as critically important for human medicine by the 5th WHO ranking (Fig. 3). While some antimicrobials were tested against all bacteria, others were only tested in certain bacteria. Particularly, not enough information was gathered to perform the meta-analyses for Campylobacter spp. and Enterococcus spp. The five groups defined in the previous paragraph were tested for susceptibility to aminoglycosides, macrolides, penicillins and tetracyclines. ‘E.coli’, ‘Salmonella’, ‘Campylobacter’ and ‘dairy’ were also tested for susceptibility to quinolones. Additionally, ‘E.coli’19,24,45 and ‘Salmonella’13,20,42 surveillance included carbapenems, 3rd and 4th generation cephalosporins, monobactams, phosphonic acid derivatives and polymyxins. Moreover, glycopeptides and oxazolidinones were tested in ‘Enterococcus’14,65,91. Three recent papers included ansamycins55,67 (S. aureus and E. coli) and glycylcyclines24 (E. coli). Finally, reports about 5th generation cephalosporins and lipopeptides, were not identified.
Qualitative synthesis of antimicrobial resistanceThis section intends to qualitatively summarize the data on AMR available in Argentina for those bacteria and antimicrobials for which there is yet not enough information to conduct a meta-analysis. It also includes data from studies based on samples from animals with clinical symptoms and isolates obtained using selective pressure or molecular selection. Therefore, it should be considered with caution.
Campylobacter spp.Five studies extracted from three articles reported prevalence of resistance within this group; one study in swine65 and four62,65,98 in chicken, with no reports from beef cattle (Fig. 3 and Suppl. Table S2).
In swine, Pantozzi et al. analyzed prevalence of resistance of Campylobacter spp. strains isolated from animals from intensive production65. Resistance to tetracycline (80%), nalidixic acid (80%), ciprofloxacin (60%), erythromycin (20%) and ampicillin (20%) was found, and absence of resistance only to gentamicin.
In chicken, Pantozzi et al. (2010) assessed one strain of Campylobacter spp., Notario et al. (2011) C. jejuni isolated from broilers and laying hen belonging to industrial and family farming, and Zbrum et al. (2015) Campylobacter spp. from hens and broilers from six poultry meat supply chains. The only strain reported by Pantozzi et al. (2010) was resistant to tetracycline and nalidixic acid and sensitive to gentamicin, ciprofloxacin, erythromycin and ampicillin65. On the other hand, Notario et al. (2011) reported that all strains isolated from industrial farming were resistant to ciprofloxacin, differing from family farming chicken where only 38.9% were resistant to this antimicrobial62. Finally, Zbrum et al. (2015) reported resistance to all antimicrobials tested including aminoglycosides, quinolones, macrolides, penicillins and tetracyclines. Among those, extremely high prevalence of AMR was reported for nalidixic acid and ciprofloxacin and very high for doxycycline and ampicillin98 (Fig. 3 and Suppl. Table S2).
Enterococcus spp.Five studies extracted from three articles reported results involving AMR of strains from the genus Enterococcus, coming from swine65,91, chicken14,65 and beef cattle65 (Fig. 3 and Suppl. Table S2).
In swine and chicken, studies on Enterococcus spp. present similar AMR patterns. Extremely high prevalence of resistance was reported for tetracycline and erythromycin, while complete susceptibility was reported for ampicillin and linezolid. With regard to vancomycin, isolates assessed by Pantozzi et al. (2010) had complete susceptibility. However, Delpech et al. (2018) and Vallejo et al. (2021) found low prevalence of resistance to vancomycin and teicoplanin in isolates from swine and chicken.
In beef cattle from extensive production, Pantozzi et al. (2010) reported moderate prevalence of resistance to tetracycline and complete susceptibility to all other antimicrobials tested, including vancomycin.
Escherichia coliE. coli was the most studied bacteria in the reports included in this review. Results from the meta-analyses for streptomycin, gentamicin, amikacin, cefotaxime, enrofloxacin, nalidixic acid, ciprofloxacin, amoxicillin with clavulanic acid, ampicillin, and tetracycline are shown in Section “Enterobacteriaceae”.
In swine, three studies assessed E. coli strains from animals with no clinical signs56,65,89, others assessed E. coli isolated from swine with diarrhea3,8,58,59, or diarrheic piglets and healthy pigs24, enterotoxicogenic E.coli35 and integron positive E.coli45 or Shiga-toxin producing E.coli12. For this food-producing animals, AMR to almost all antimicrobial agents evaluated was observed, except for carbapenems and glycylcyclines. Isolates assessed in the initial reports were susceptible to colistin56,57,65,66 until 2019; Faccone et al. (2019) reported extremely high levels of AMR to cefotaxime and high levels of AMR to colistin24, when isolating E. coli using plates containing cefotaxime, colistin or both. Moreover, at least one study reported very high resistance to enrofloxacin3,8,45 and ciprofloxacin24,58. In swine with diarrhea, extremely high prevalence of AMR to erythromycin, tilmicosin and tylosin was found3,8.
In chicken, Tessi et al. (1997), Pantozzi et al. (2010) and Dominguez et al. (2017) studied E. coli, while Dominguez et al. (2018) studied colistin-resistant E. coli, most of them from intensive farming18,19,65 or not described86. With respect to colistin, 49% of E. coli strains were resistant to this antimicrobial18. Furthermore, colistin-resistant E. coli testing showed higher prevalence of resistance than E. coli to all antimicrobials except amikacin19. Thus, extremely high prevalence of AMR was reported for streptomycin, ceftiofur, cefotaxime, cefepime, enrofloxacin, nalidixic acid, ampicillin and tetracycline19. Finally, complete susceptibility to imipenem and meropenem was reported19 (Fig. 3 and Suppl. Table S2).
In beef cattle, Pantozzi et al. (2010) and Torres et al. (2017) assessed E. coli strains in healthy animals raised in extensive production65,89. Absence of AMR was reported for ceftiofur89 (Fig. 3). In E.coli79 and Shiga toxin-producing E.coli12,44,52 from diarrheic dairy and beef-cattle calves, authors reported high resistance to polymyxin B52, very high resistance to gentamicin52, amoxicillin/clavulanic acid79 and extremely high AMR to neomycin52,79, enrofloxacin79 and tetracycline52 (Fig. 3 and Suppl. Table S2).
In dairy, studies assessed E. coli from healthy cows or diarrheic calves. Torres et al. (2017) assessed E. coli isolated from vaginal samples of healthy cows. The authors reported complete susceptibility to ceftiofur. Results from healthy cows (see Section “Enterobacteriaceae”) contrast with those obtained when assessing E.coli79, enterotoxicogenic E.coli67, and Shiga-toxin producing E.coli44 from diarrheic calves. Prevalence of AMR in diarrheic calves ranged from rare to extremely high for streptomycin, kanamycin, and amoxicillin with clavulanic acid. Pasayo et al. (2019) reported resistance to rifampicin in the three isolates assessed67, while most authors reported complete susceptibility to cefotaxime (Fig. 3 and Suppl. Table S2).
Salmonella spp.Eleven studies reported prevalence of AMR within this group; seven studies in swine13,39,42,66,92,93, three in chicken20,72,86 and one in beef cattle65 (Fig. 3 and Suppl. Table S2). Results from the meta-analyses for streptomycin, gentamicin, amikacin, cefotaxime, enrofloxacin, nalidixic acid, ciprofloxacin, amoxicillin with clavulanic acid, ampicillin, and tetracycline are shown in Section “Enterobacteriaceae”.
In swine, most studies evaluated AMR patterns in Salmonella spp. stains isolated from intensive production, including confined farrow-to-finish farms13,66,92,93, while Joaquim et al. (2021) compared intensive production to backyard farms. Prevalence of AMR was rare to fosfomycin93 and polymyxins92,93. The most recent study (2021) showed low resistance to 3rd and 4th generation cephalosporins, ceftiofur and cefepime, in intensive production42. Extremely high AMR was reported for amoxicillin92 (Fig. 3 and Suppl. Table S2).
In chicken, Tessi et al. (1997) and Rodríguez et al. (2018) assessed Salmonella spp. while Dominguez et al. (2021) assessed Salmonella Heidelberg. On the one hand, all strains isolated from 657 tested-chicken from family farms were susceptible to all antimicrobials tested, including phosphonic acid derivatives72. On the other hand, when assessing Salmonella Heidelberg from intensive production, very high prevalence of AMR was reported for ceftiofur and cefepime20.
Mastitis-causing bacteriaIn dairy, the prevalence of AMR was tested for a lower number of antimicrobials. For S. aureus, all authors tested AMR to penicillin and erythromycin and results from the meta-analyses are presented in Section “Staphylococcus aureus”. The main fluoroquinolone tested was enrofloxacin and results showed rare prevalence of resistance for S.aureus55,73. Other bacteria assessed were coagulase-negative staphylococci32,71,80,82,83, coagulase-positive staphylococci11 and Streptococcus spp.16,36 (Fig. 2). The highest AMR prevalence reported in mastitis-causing bacteria was 83.3% of kanamycin-resistant Streptococcusagalactiae36.
MDR was low68,81 or absent16,55,73 in Staphylococcus spp. isolated from dairy, with the highest value being 5.9% reported for coagulase-negative staphylococci71. However, MDR was considerably higher in S. agalactiae, where 25% of the isolates were resistant to three or more antimicrobial clases36.
Quantitative synthesis of antimicrobial resistance: meta-analysesTwenty-eight studies, of sixty-five included in the qualitative synthesis, were finally included in the quantitative synthesis (Suppl. Fig. S1). Among food-transmitted bacteria, twenty-one studies involved Enterobacteriaceae, while not enough studies were available to perform meta-analyses for Campylobacter spp. or Enterococcus spp. Among mastitis-causing bacteria, there were enough studies only for S. aureus.
EnterobacteriaceaeThe number of studies that analyzed the AMR of isolates depended on the antimicrobial agent considered. At least ten studies were available for streptomycin, gentamicin, amikacin, cefotaxime, enrofloxacin, nalidixic acid, ciprofloxacin, amoxicillin with clavulanic acid, ampicillin, and tetracycline. The pooled estimate of the prevalence of Enterobacteriaceae bacteria resistant to each of these antimicrobials is shown in Table 1. The highest prevalence of AMR was observed for tetracycline, followed by ampicillin, streptomycin and nalidixic acid. The lowest prevalence was observed for gentamicin and amikacin. Between-study heterogeneity was substantial for most antimicrobials. Funnel plots estimating publication bias are shown in Suppl. Fig. S1. According to the Egger's regression test, most antimicrobials could be affected by publication bias related to small studies, except for ampicillin, tetracycline, nalidixic acid and MDR (Table 1).
Prevalence of multidrug resistance in bacteria isolated from food-producing animals in Argentina.
| Antimicrobial | No. of studies | No. of isolates | Pooled estimatea | 95% CI | I2b | Cochran Qp-value | Egger'sp-valuec |
|---|---|---|---|---|---|---|---|
| Enterobacteriaceae | |||||||
| Streptomycin | 17 | 646 | 0.2659 | [0.1632; 0.4022] | 82.0% | <0.0001 | 0.0345 |
| Gentamicin | 17 | 700 | 0.0753 | [0.0328; 0.1635] | 71.9% | <0.0001 | 0.0017 |
| Amikacin | 10 | 380 | 0.0649 | [0.0196; 0.1939] | 80.4% | <0.0001 | 0.0219 |
| Amikacin* | 9 | 331 | 0.0483 | [0.0181; 0.1229] | 31.9% | 0.1629 | NA |
| Cefotaxime | 11 | 440 | 0.0327 | [0.0064; 0.1516] | 77.6% | <0.0001 | 0.0006 |
| Cefotaxime* | 10 | 420 | 0.0154 | [0.0075; 0.0314] | 0.0% | 0.8745 | 0.0003 |
| Enrofloxacin | 11 | 390 | 0.1006 | [0.0405; 0.2285] | 76.4% | <0.0001 | 0.0177 |
| Nalidixic acid | 16 | 688 | 0.2425 | [0.1103; 0.4527] | 86.0% | <0.0001 | 0.0501 |
| Ciprofloxacin | 12 | 430 | 0.0890 | [0.0380; 0.1944] | 59.8% | 0.0041 | 0.0188 |
| Amoxicillin/clav. acid | 14 | 445 | 0.0752 | [0.0248; 0.2061] | 79.2% | <0.0001 | 0.0243 |
| Ampicillin | 19 | 685 | 0.3275 | [0.1880; 0.5061] | 86.2% | <0.0001 | 0.3172 |
| Tetracycline | 20 | 697 | 0.4701 | [0.2941; 0.6538] | 87.7 | <0.0001 | 0.3631 |
| Multidrug resistance | 0.6312 | ||||||
| Staphylococcus aureus | |||||||
| Penicillin | 7 | 663 | 0.2556 | [0.1412; 0.4177] | 84.6% | <0.0001 | NA |
| Erythromycin | 6 | 633 | 0.1245 | [0.0365; 0.3484] | 85.5% | <0.0001 | NA |
Meta-analysis excluding outliers or influential studies: amikacin excluding Pantozzi et al. (2010) cattle; cefotaxime excluding Dominguez et al. (2021). (a) Pooled estimate prevalence of isolates resistant to the antimicrobials. (b) Heterogeneity index. (c) Egger's regression test of Funnel plot asymmetry. CI: confidence interval; NA: not available due to study limitations.
The AMR pattern of streptomycin, ampicillin and tetracycline was associated with the animal species from which they were isolated (p<0.01) (Table 2). Enterobacteriaceae isolates from swine (followed by chicken) showed the highest prevalence of resistance to the three antimicrobials, with pooled estimates ranging from 0.40 [95% CI=0.22; 0.63] for streptomycin to 0.64 [95% CI=0.39; 0.83] for tetracycline. On the contrary, isolates from beef-cattle showed the lowest prevalence of resistance to the same antimicrobials, with pooled estimates ranging from 0.07 [95% CI=0.01; 0.42] for streptomycin to 0.10 [95% CI=0.04; 0.21] for tetracycline.
Subgroup meta-analyses for Enterobacteriaceae from food-producing animals, by stratification variable.
| Antimicrobial | Subgroup | N̊ studies | Pooled estimate | 95% CI | Q | I2 | Qs | p-values | |
|---|---|---|---|---|---|---|---|---|---|
| Streptomycin | |||||||||
| Animal | Swine | 8 | 0.4045 | [0.2161; 0.6260] | 44.39 | 84.2% | 12.17 | 0.0068 | |
| Chicken | 5 | 0.2770 | [0.1232; 0.5110] | 11.05 | 63.8% | ||||
| Cattle | 3 | 0.0659 | [0.0069; 0.4191] | 2.00 | 0.0% | ||||
| Dairy | 1 | 0.1667 | [0.0299; 0.5651] | 0.00 | -- | ||||
| System | Nd | 4 | 0.4197 | [0.1618; 0.7304] | 32.17 | 90.7% | 8.40 | 0.0150 | |
| Intensive | 6 | 0.3393 | [0.1737; 0.5565] | 20.97 | 76.2% | ||||
| Extensive | 7 | 0.1139 | [0.0392; 0.2883] | 15.25 | 60.7% | ||||
| Bacteria | E. coli | 10 | 0.2990 | [0.1452; 0.5185] | 70.05 | 87.2% | 0.88 | 0.3494 | |
| Salmonella | 7 | 0.2008 | [0.0872; 0.3980] | 11.05 | 45.7% | ||||
| Gentamicin | |||||||||
| Animal | Swine | 10 | 0.0999 | [0.0314; 0.2755] | 35.42 | 74.6% | 1.08 | 0.5833 | |
| Chicken | 5 | 0.0427 | [0.0049; 0.2860] | 17.05 | 76.5% | ||||
| Cattle | 2 | 0.0420 | [0.0000; 1.0000] | 1.90 | 47.5% | ||||
| System | Nd | 4 | 0.0224 | [0.0012; 0.3041] | 13.21 | 77.3% | 4.31 | 0.1157 | |
| Intensive | 8 | 0.1429 | [0.0428; 0.3833] | 26.55 | 73.6% | ||||
| Extensive | 5 | 0.0480 | [0.0124; 0.1685] | 2.68 | 0.0% | ||||
| Bacteria | E. coli | 6 | 0.0594 | [0.0136; 0.2246] | 17.80 | 71.9% | 0.22 | 0.6423 | |
| Salmonella | 11 | 0.0850 | [0.0248; 0.2531] | 36.40 | 72.5% | ||||
| Amikacin | |||||||||
| Animal | Swine | 6 | 0.0322 | [0.0054; 0.1698] | 8.72 | 42.7% | 3.14 | 0.2084 | |
| Chicken | 2 | 0.2002 | [0.0000; 1.0000] | 5.24 | 80.9% | ||||
| Cattle | 2 | 0.1085 | [0.0018; 0.8913] | 0.19 | 0.0% | ||||
| System | Nd | 2 | 0.0199 | [0.0000; 0.9999] | 1.94 | 48.5% | 1.70 | 0.4266 | |
| Intensive | 5 | 0.0942 | [0.0065; 0.6236] | 19.65 | 79.6% | ||||
| Extensive | 3 | 0.0771 | [0.0092; 0.4287] | 0.93 | 0.0% | ||||
| Bacteria | E. coli | 5 | 0.0700 | [0.0092; 0.3792] | 32.89 | 87.8% | 0.04 | 0.8326 | |
| Salmonella | 5 | 0.0553 | [0.0046; 0.4271] | 8.13 | 50.8% | ||||
| Cefotaxime | |||||||||
| Animal | Swine | 7 | 0.0157 | [0.0052; 0.0463] | 3.93 | 0.0% | 1.37 | 0.5043 | |
| Chicken | 3 | 0.2071 | [0.0000; 0.9999] | 23.12 | 91.4% | ||||
| Cattle | 1 | 0.0109 | [0.0001; 0.5725] | 0.00 | -- | ||||
| System | Nd | 2 | 0.0062 | [0.0009; 0.0392] | 0.02 | 0.0% | 13.14 | 0.0014 | |
| Intensive | 6 | 0.0659 | [0.0025; 0.6632] | 35.84 | 86.0% | ||||
| Extensive | 3 | 0.0227 | [0.0044; 0.1094] | 0.44 | 0.0% | ||||
| Bacteria | E. coli | 5 | 0.0101 | [0.0078; 0.0131] | 0.09 | 0.0% | 2.96 | 0.0854 | |
| Salmonella | 6 | 0.0841 | [0.0035; 0.7070] | 33.64 | 85.1% | ||||
| Enrofloxacin | |||||||||
| Animal | Swine | 8 | 0.1148 | [0.0312; 0.3431] | 34.09 | 79.5% | 0.56 | 0.9061 | |
| Chicken | 1 | 0.0385 | [0.0008; 0.6604] | 0.00 | -- | ||||
| Cattle | 1 | 0.0652 | [0.0038; 0.5614] | 0.00 | -- | ||||
| Dairy | 1 | 0.0556 | [0.0021; 0.6243] | 0.00 | -- | ||||
| System | Nd | 2 | 0.0507 | [0.0000; 1.0000] | 5.20 | 80.8% | 1.87 | 0.3931 | |
| Intensive | 3 | 0.2197 | [0.0060; 0.9296] | 13.12 | 84.8% | ||||
| Extensive | 6 | 0.0707 | [0.0238; 0.1919] | 7.07 | 29.3% | ||||
| Bacteria | E. coli | 5 | 0.1041 | [0.0283; 0.3166] | 7.39 | 45.9% | 0.02 | 0.8762 | |
| Salmonella | 6 | 0.0917 | [0.0145; 0.4086] | 28.24 | 82.3% | ||||
| Nalidixic acid | |||||||||
| Animal | Swine | 10 | 0.2883 | [0.1414; 0.4992] | 45.66 | 80.3% | 4.52 | 0.1043 | |
| Chicken | 4 | 0.2006 | [0.0013; 0.9800] | 32.67 | 90.8% | ||||
| Cattle | 2 | 0.0947 | [0.0002; 0.9830] | 0.38 | 0.0% | ||||
| System | Nd | 4 | 0.0584 | [0.0018; 0.6837] | 37.00 | 91.9% | 21.67 | <0.0001 | |
| Intensive | 8 | 0.4846 | [0.3034; 0.6698] | 35.85 | 80.5% | ||||
| Extensive | 4 | 0.1204 | [0.0516; 0.2559] | 1.74 | 0.0% | ||||
| Bacteria | E. coli | 6 | 0.2253 | [0.0414; 0.6622] | 45.68 | 89.1% | 0.02 | 0.8858 | |
| Salmonella | 10 | 0.2501 | [0.0797; 0.5622] | 61.57 | 85.4% | ||||
| Ciprofloxacin | |||||||||
| Animal | Swine | 8 | 0.0970 | [0.0275; 0.2899] | 21.44 | 67.4% | 0.38 | 0.8264 | |
| Chicken | 2 | 0.0765 | [0.0002; 0.9735] | 0.47 | 0.0% | ||||
| Cattle | 2 | 0.0415 | [0.0000; 1.0000] | 1.90 | 47.5% | ||||
| System | Nd | 1 | 0.0053 | [0.0002; 0.1041] | 0.00 | -- | 5.15 | 0.0763 | |
| Intensive | 7 | 0.1325 | [0.0497; 0.3085] | 16.03 | 62.6% | ||||
| Extensive | 4 | 0.0557 | [0.0085; 0.2896] | 2.45 | 0.0% | ||||
| Bacteria | E. coli | 4 | 0.0576 | [0.0116; 0.2416] | 4.29 | 30.0% | 1.06 | 0.3043 | |
| Salmonella | 8 | 0.1186 | [0.0341; 0.3391] | 16.53 | 57.6% | ||||
| Amoxicillin/clav. acid | |||||||||
| Animal | Swine | 8 | 0.0810 | [0.0198; 0.2781] | 19.22 | 63.6% | 1.53 | 0.6764 | |
| Chicken | 2 | 0.2412 | [0.0000; 1.0000] | 14.18 | 92.9% | ||||
| Cattle | 3 | 0.0267 | [0.0004; 0.6288] | 2.49 | 19.6% | ||||
| Dairy | 1 | 0.0263 | [0.0003; 0.6893] | 0.00 | -- | ||||
| System | Nd | 1 | 0.0071 | [0.0001; 0.3312] | 0.00 | -- | 2.84 | 0.2412 | |
| Intensive | 8 | 0.1328 | [0.0243; 0.4845] | 43.55 | 83.9& | ||||
| Extensive | 5 | 0.0395 | [0.0076; 0.1802] | 4.74 | 15.6% | ||||
| Bacteria | E. coli | 9 | 0.0357 | [0.0109; 0.1105] | 19.25 | 58.4% | 4.99 | 0.0255 | |
| Salmonella | 5 | 0.2669 | [0.0308; 0.8067] | 23.81 | 83.2% | ||||
| Ampicillin | |||||||||
| Animal | Swine | 11 | 0.4345 | [0.2438; 0.6469] | 45.60 | 78.1% | 21.35 | <0.0001 | |
| Chicken | 4 | 0.3771 | [0.0385; 0.9015] | 16.12 | 81.4% | ||||
| Cattle | 3 | 0.0672 | [0.0149; 0.2550] | 0.75 | 0.0% | ||||
| Dairy | 1 | 0.1111 | [0.0099; 0.6105] | 0.00 | -- | ||||
| System | Nd | 3 | 0.3900 | [0.0253; 0.9404] | 46.04 | 95.7% | 13.24 | 0.0013 | |
| Intensive | 9 | 0.5050 | [0.2636; 0.7441] | 38.58 | 79.3% | ||||
| Extensive | 7 | 0.1134 | [0.0482; 0.2440] | 11.91 | 49.6% | ||||
| Bacteria | E. coli | 10 | 0.2545 | [0.1110; 0.4828] | 86.32 | 89.6% | 1.37 | 0.2422 | |
| Salmonella | 9 | 0.4399 | [0.1787; 0.7392] | 42.43 | 81.1% | ||||
| Tetracycline | |||||||||
| Animal | Swine | 11 | 0.6362 | [0.3896; 0.8274] | 66.13 | 84.9% | 36.32 | <0.0001 | |
| Chicken | 5 | 0.4180 | [0.1095; 0.8076] | 22.18 | 82.0% | ||||
| Cattle | 3 | 0.0993 | [0.0445; 0.2071] | 0.21 | 0.0% | ||||
| Dairy | 1 | 0.3333 | [0.0365; 0.8685] | 0.00 | -- | ||||
| System | Nd | 3 | 0.6887 | [0.0624; 0.9866] | 26.24 | 92.4% | 24.93 | <0.0001 | |
| Intensive | 9 | 0.6721 | [0.4376; 0.8438] | 38.91 | 79.4% | ||||
| Extensive | 8 | 0.1680 | [0.0940; 0.2819] | 10.56 | 33.7% | ||||
| Bacteria | E. coli | 10 | 0.5430 | [0.2348; 0.8215] | 119.37 | 92.5% | 0.76 | 0.3820 | |
| Salmonella | 10 | 0.3880 | [0.2049; 0.6093] | 26.92 | 66.6% | ||||
| Multidrug resistance | |||||||||
| Animal | Swine | 11 | 0.4733 | [0.2909; 0.6631] | 66.54 | 85.0% | 12.57 | 0.0057 | |
| Chicken | 4 | 0.3840 | [0.0194; 0.9516] | 13.36 | 77.5% | ||||
| Cattle | 2 | 0.0682 | [0.0000; 0.9979] | 0.75 | 0.0% | ||||
| Dairy | 1 | 0.0556 | [0.0031; 0.5235] | 0.00 | -- | ||||
| System | Nd | 4 | 0.3671 | [0.1530; 0.6507] | 26.77 | 88.8% | 19.82 | <0.0001 | |
| Intensive | 8 | 0.6191 | [0.3439; 0.8345] | 29.69 | 76.4% | ||||
| Extensive | 6 | 0.1066 | [0.0432; 0.2399] | 6.05 | 17.4% | ||||
| Bacteria | E. coli | 7 | 0.2916 | [0.1025; 0.5974] | 37.5 | 84.0% | 0.76 | 0.3838 | |
| Salmonella | 11 | 0.4415 | [0.1935; 0.7225] | 77.27 | 87.1% | ||||
(s) Cochran's Q equivalent and test of between-study heterogeneity for subgroups. CI, confidence interval. Significant associations among stratification variables (Animal, System, Bacteria) and AMR are indicated in bold letters.
The AMR pattern of Enterobacteriaceae to streptomycin, cefotaxime, nalidixic acid, ampicillin and tetracycline was associated with the animal production system from which they were isolated (p<0.05) (Table 2). Isolates coming from intensive production showed higher prevalence of resistance to each antimicrobial than isolates from extensive production. In intensive production, pooled estimates for these antimicrobials ranged from 0.066 [95% CI=0.002; 0.663] for cefotaxime to 0.67 [95% CI=0.438; 0.844] for tetracycline. In extensive animal production, pooled estimates for these antimicrobials ranged from 0.023 [95% CI=0.004; 0.109] for cefotaxime to 0.17 [95% CI=0.094; 0.282] for tetracycline.
Prevalence of AMR to amoxicillin with clavulanic acid was the only one associated with the bacteria within Enterobacteriaceae (p<0.05) (Table 2). In this case, isolates from the genus Salmonella had a higher pooled estimate of AMR than E. coli,i.e., 0.27 and 0.04 respectively.
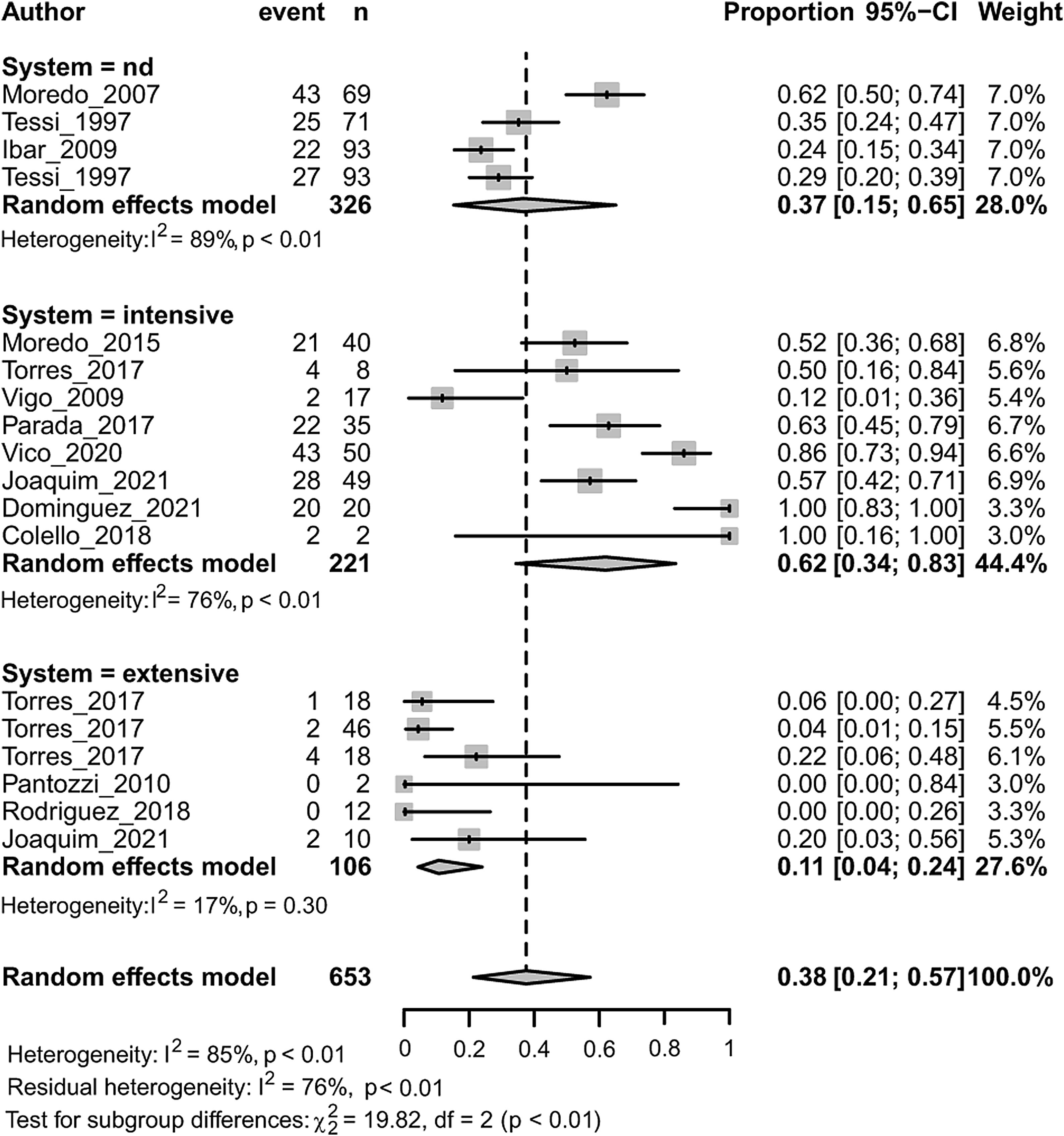
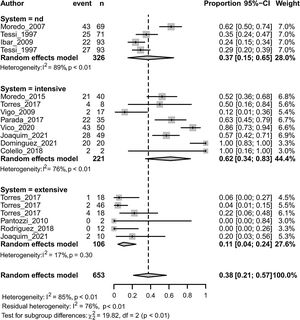
MDR in Enterobacteriaceae was associated with both the animal species (p<0.01) and the animal production system (p<0.001) (Fig. 4). Within each subgroup analysis, swine (followed by chicken) and intensive production showed the highest prevalence of MDR with respective pooled estimates of 0.47 [0.29; 0.66] and 0.62 [0.34; 0.83]. Dairy (followed by beef-cattle) and extensive production showed the lowest prevalence of MDR with respective pooled estimates of 0.056 [0.003; 0.524] and 0.107 [0.043; 0.240]. Results on dairy production should be considered with caution because there is only one study included.
Staphylococcus aureusAt least six studies were available with results on the prevalence of resistance to penicillin and erythromycin. The pooled estimates of the prevalence of S. aureus resistant to each of these antimicrobials is shown in Table 1. The highest prevalence of AMR was observed for penicillin. Between-study heterogeneity was substantial for both antimicrobials. Funnel plots estimating publication bias showed some degree of asymmetry (Suppl. Fig. S2).
DiscussionWithin the antimicrobials authorized by SENASA for their use in animals, 85% are shared with human medicine or belong to groups classified by the WHO as critically important antimicrobials for humans (SENASA Resolution 559/2011)97. Given that the use of antimicrobials in animals increases AMR in humans37, there is enough justification to manage their use, fate and AMR surveillance in food-producing animals and the open environment. The study of AMR in animal production in Argentina, as in many other countries, is not yet completely coordinated nor harmonized and there are still important gaps. The type of samples and antimicrobials tested differed widely among authors. However, most authors agreed with the methods and guidelines followed to interpret AMR. The CLSI guidelines were the ones preferred in Argentina, together with the EUCAST guidelines, which are both the most popular worldwide43.
Meta-analyses are a powerful tool to quantitatively summarize available data and assess possible sources of heterogeneity, such as animal species or production systems35. Further studies are needed, particularly for Campylobacter spp. and Enterococcus spp., because not enough data is yet available to perform meta-analyses. The same situation occurs for some classes of antimicrobials classified as critically important for human medicine by the 5th report from the WHO, including polymyxins and glycopeptides.
In this study, the AMR profiles from the qualitative synthesis could be divided into two main categories: AMR developed to antimicrobials authorized for veterinary use (SENASA Resolution 559/2011) and AMR developed to antimicrobials used exclusively for human medicine97. The first category includes the combination of amoxicillin and clavulanic acid, ampicillin, ceftiofur, doxycycline, enrofloxacin, erythromycin, fosfomycin, gentamicin, kanamycin, neomycin, penicillin, rifampicin, spiramycin, streptomycin, tetracycline, tilmicosin and tylosin. The second category includes amikacin, aztreonam, cefepime, cefotaxime, ciprofloxacin, colistin (since 2019 banned for animal use), nalidixic acid and polymyxin B. Although all of them are critically important antimicrobials, the second category is the most alarming in terms of public health.
According to the WHO ranking, the following are ‘highest priority critically important antimicrobials’: quinolones, 3rd-generation cephalosporins or higher, macrolides and ketolides, glycopeptides and polymyxins97. In the following paragraphs some results of these antimicrobials from the qualitative synthesis and meta-analyses performed, will be discussed.
QuinolonesQuinolones are one of the only therapies available to treat severe Salmonella spp. and E. coli infections in humans97. In this study, the reports included in the qualitative synthesis showed prevalence of resistance to quinolones ranging from extremely high in Campylobacter spp. from both swine65, and chicken62,98, in E. coli from chicken19 and diarrheic beef-cattle calves79, and very high in Salmonella spp. isolated from swine66 and chicken20. These results are similar to those from countries where the use of fluoroquinolones is approved in food-producing animals, such as E. coli isolated from chickens in China99 and reports from the EU21. When the pooled prevalence of AMR to quinolones was estimated for Enterobacteriaceae, the levels found were low, moderate, and high for ciprofloxacin, enrofloxacin and nalidixic acid, respectively. In Australia, a country where fluoroquinolones are preserved, complete susceptibility to ciprofloxacin was reported in commensal E. coli and Salmonella spp. isolated from meat chickens1.
Moreover, in human medicine, the Latin American Network for Antimicrobial Resistance Surveillance reported 70.4% ciprofloxacin-resistant Campylobacter spp. in Argentina46, representing a serious threat to public health. On the contrary, ciprofloxacin-resistant Campylobacter spp. is rare or low in countries where the use of fluoroquinolones has never been approved in food-producing animals90.
In Argentina, ciprofloxacin is exclusively used for human medicine; however, animal administration of enrofloxacin can lead to similar concentrations of plasmatic enrofloxacin and ciprofloxacin4,15,29. Enrofloxacin was not on the list of approved growth promoters submitted to the OIE in 2013 and 2014, therefore the extremely high levels of resistance observed could be related to misuse. Concordantly, residual levels of enrofloxacin considered toxic for human consumption were recently found in dehydrated eggs from Argentina85.
CephalosporinsIn Argentina, the main cephalosporins available for veterinary use are ceftiofur and cefquinome (SENASA Resolution 559/2011). In the qualitative synthesis of this study, data on resistance to ceftiofur come from E.coli3,19,89 and Salmonella spp.42,92. Prevalence of resistance in swine is in between what has been reported in US (very low) and China (moderate)26,99, or higher3,24. For E. coli from cattle, susceptibility to cefotaxime was complete in animals at local slaughterhouses, while high prevalence of cefotaxime-resistance has been associated with diarrheic beef and dairy calves in Argentina and grazing beef cattle in the USA (Florida)65,89. When the pooled prevalence of AMR to cefotaxime was estimated for Enterobacteriaceae, the levels found were low. Therefore, local resistance to 3rd generation cephalosporins seems to be related to animals with clinical symptoms.
Finally, the extremely high prevalence of resistance to cephalosporins reported for colistin-resistant E. coli from chicken could be a result of co-selection due to the administration of colistin as growth promoter41.
MacrolidesIn this study, up to an extremely high prevalence of macrolide resistance was observed in swine and chicken, including Salmonella spp. from family-farming chicken. Resistance to macrolides in Enterococcus spp.65 from swine and chicken was higher than that reported in the USA. However, resistance to erythromycin in US beef cattle was moderate while in this study susceptibility was complete26. The pooled prevalence of erythromycin resistance in S. aureus from dairy cattle estimated in this study (0.125) was higher than the worldwide one (0.085) estimated by Molineri et al.54.
In Argentina, tylosin is administered as a growth promoter (Luna F., personal communication, 2022, SENASA). This use is most likely contributing to the selection of macrolide-resistant bacteria10. Moreover, another non-therapeutic use of macrolides is as metaphylactics in calves entering feedlots to prevent bovine respiratory disease25. Therefore, it would be of great importance to know the levels of resistance in this type of production system.
TetracyclinesAs Argentina still has endemic Brucella spp. infections, the recommendation is to continue considering tetracyclines as critically important antimicrobials97. Despite this recommendation, chlortetracycline and oxytetracycline are administered as growth promoters in food-producing animals (Luna F., personal communication, 2022, SENASA). In the qualitative synthesis, resistance to tetracycline increases over time until reaching extremely high levels of resistance in E. coli, Enterococcus spp. and Campylobacter spp. (except bacteria from beef and dairy cattle). Similar results were also reported in the USA, China and the EU21,26,99.
As described in some global meta-analyses75,76, in our study, tetracycline was the antimicrobial with the highest pooled prevalence of AMR in Enterobacteriaceae from Argentina. Particularly in intensive production, AMR was similar to the pooled prevalence reported for Salmonella spp. from swine in China (0.68, 95% CI: 0.59, 0.77)76.
Extremely high levels of resistance to tetracyclines do not only represent loss of the effectiveness of these antimicrobials, but it has also been suggested that tetracycline-resistant E. coli is more likely to be resistant to other antimicrobials74.
PolymyxinsIn Argentina, colistin was the only polymyxin authorized in animals and was extensively used as a growth promoter. Resistance to colistin can occur by chromosomal mutations and by acquisition of plasmid-carrying determinants, mainly mcr-1. Quiroga et al. (2019) have reviewed the outburst of this resistance gene in Latin America and the risk of the rapid dissemination of mcr-1 has alerted actors worldwide70.
In Argentina, the alarming results reported by independent researchers and the national surveillance program, have led to new regulations. In line with the WHO and OIE recommendations and to preserve the efficacy of colistin, on January 2019 SENASA banned the production, distribution, import, use and possession of veterinary products containing colistin (RESOL-2019-22-APN-PRES#SENASA).
To date, there is not enough available information about this group of antimicrobials to perform meta-analyses, which will be necessary to estimate the effect of the intervention applied in the country. Of special importance is the methodology followed to test AMR to colistin, given that disk-diffusion is considered unreliable because colistin diffuses poorly into agar, resulting in smaller inhibition zones47.
GlycopeptidesGlycopeptides were never authorized for their use in Argentine animal production. Concordantly, vancomycin-resistant Enterococcus spp.65 was neither isolated from swine, chicken nor beef cattle, until recently. Since 2018, information about glycopeptide resistance has been reported for swine and chicken14,91. Resistance to this antimicrobial is common in countries where avoparcin was used as feed additive22. Similarly, as occurs with colistin, there are not enough data available on this group to perform meta-analyses.
AMR, animal species and animal production systemsIndustrialization of agriculture has been accompanied by important benefits and risks to human, animal, and environmental health. In many developing countries, the change occurred at such a high speed that caused social impacts with increased risk of marginalization of smallholder farmers33. When analyzing the transitions in food animal production from the One Health approach, Silbergeld et al. (2019) states that the use of antimicrobial drugs in food animal production is the innovation that has most profoundly and dangerously transformed human health at the global level. In concordance, the author calls for an end to the use of antimicrobials for non-therapeutic purposes and the cruelties of confinement78. In our study, a reduced number of the articles included compared AMR among different animal species65 or production systems42,62,89. However, through meta-analyses of proportions followed by subgroup analyses, it was possible to test the association between AMR and the variables ‘animal species’ and ‘animal production systems’35, as previously applied in global meta-analyses77,84. Patterns of AMR to streptomycin, ampicillin, tetracycline and MDR were associated with the animal species, while patterns of AMR to streptomycin, cefotaxime, nalidixic acid, ampicillin, tetracycline and MDR were associated with the animal production system from which they were isolated (Table 2 and Fig. 4). Greater prevalence of resistance was associated with swine (followed by chicken), and with intensive production. On the contrary, beef (or dairy) cattle and, extensive production, showed the lowest prevalence of resistance and resistance to a lesser number of antimicrobials.
Food-producing animals are not only vehicles of AMR transmission but also endpoints in the dissemination, selection, and spread of antimicrobial-resistant bacteria and AMR genes87. Among the studies included in this review, authors reported the presence of a variety of resistance-encoding genes and cassettes including, aac(6′)-Ie-aph(2′)-Ia14, aadA144, aadA567, aphA336, blaCTX-M-1/1524, blaCTX-M-219,24, blaCTX-M-8/2524, blaCTX-M-9/1419,24, blaCMY19,24, blaPER-224, blaTEM44, blaZ55, dhfrA1767, drfA144, ermB36, ermC55, floR44, lnu(B)36, mcr-118,19,24, mecA80, oqxAB19, qepA19, qnrA19, qnrB19, qnrD19, qnrS19, strA, strB, sul1, sul2, tet(A),tet(B44), tet(M)36, tet(O)36, vanA14. Therefore, a seemingly beneficial practice that would be the redirection of livestock manure to soils to close the cycles of nutrients and the organic matter now represents an ideal scenario for the dissemination of these contaminants. Thus, manure is a very difficult waste to dispose of, which in many cases leads to its accumulation in small areas with subsequent contamination of soil, air and ground water.
Due to the expansion and globalization of food and animal trade, international tourism and host movements, AMR can easily surpass local, national and continental borders, making AMR a worldwide problem51. In the EU and North America surveillance data are available and research networks are active; however, these are still largely lacking in low and middle income countries94. Moreover, it has been stated that AMR is aggravated in developing countries due to the gross abuse in the use of antimicrobials, including non-human uses5. In between, there are those countries, with a long history of extensive systems, such as Argentina, which in the last decades have started to shift gradually to intensive, industrialized, or confined systems, and to increase chicken and swine production. Limited interventions in one country, continent and for instance, in the developing world, could compromise the efficacy and endanger the AMR containment policies implemented in other parts of the world28.
Answering to the need for global surveillance programs, the OIE encourages the application of national surveillance programs, whose results are being integrated in global reports64. This approach will provide guidance for antimicrobial use and tools to detect and contain the development and spread of AMR worldwide.
ConclusionsThe data presented in this study gives documented evidence that support the hypothesis that AMR to common food-transmitted bacteria is reaching alarming levels in our country. This situation was especially observed in animal production systems related to confined-intensive breeding relying on the habitual use of antimicrobials as is mostly the case for swine and chicken production in Argentina. In addition, we identified that there are still important local gaps of information regarding beef-cattle from feedlot, as well as Campylobacter spp. and Enterococcus spp. All these findings emphasize the urgent need for National coordination. Restrictive measures for non-therapeutic and/or massive uses of highest priority antimicrobials should be applied to maintain the efficacy of these chemical agents, to preserve human and animal health as well as to ensure food safety and food security.
FundingThis work was supported by UBACYT [20020190200419BA] and LOMASCyT [FCA-91 Res. N° 857/2018], Argentina. The funding sources were not involved in, the study design, the collection, analysis and interpretation of data, the writing of the report, nor the decision to submit the article for publication.
Conflict of interestNone declared.