Diverse habitats have been screened for novel antimicrobial actinomycetes, while others remain unexplored. In this study, we analyzed the bioactivities of actinomycetes cultured from rhizosphere soils of the desert plant Artemisia tridentata and the nearby bulk soils. Actinomycetes were screened for antifungal and antibacterial activities toward a panel of plant pathogens; all comparisons were between activities of rhizosphere soil isolates toward those of its counterpart bulk soil. A selected group of the strongest antifungal isolates were also tested against two antifungal-drug resistant strains of Candida albicans. 16S rDNA partial sequences and phylogenetic analysis of isolates that showed broad-spectrum antifungal activities were performed. Forty-two out of 200 and two soil isolated actinomycetes were selected for their strong antifungal activities. The highest proportion of isolates (p<0.05) from rhizosphere soil of an old plant showed antagonism against gram-positive bacteria (0.483 and 0.224 proportions against Bacillus subtilis and Rathayibacter tritici, respectively), and phytopathogenic fungi (0.259, 0.431, and 0.345 proportions against Fusarium oxysporum, Rhizoctonia solani and Pythium ultimum, respectively), while the highest antagonism against the gram-negative bacteria predominated in isolates from the bulk soils. Isolates from a rhizosphere soil of a young plant were characterized for strong antagonist activities against Fusarium oxysporum (0.333 proportion, p<0.05). Phylogenetic analysis of 16S rDNA sequences showed that isolates that exhibited strong antifungal activity were genetically similar. We conclude that the rhizosphere soil of A. tridentata is an excellent source for discovery of actinomycetes with potentially novel antifungal compounds.
En la búsqueda de actinomicetos antimicrobianos se han estudiado diversos hábitats, pero muchos permanecen aún sin explorar. En este estudio analizamos las actividades biológicas de cultivos de actinomicetos provenientes de suelos rizosféricos de la planta desértica Artemisiatridentata y de suelos no asociados a sus raíces. Los actinomicetos fueron seleccionados por sus actividades antifúngicas y antibacterianas contra un panel de patógenos de plantas. Todas las comparaciones fueron entre las actividades de los aislados rizosféricos y aquellas de los aislados no asociados a las raíces. Un grupo selecto de los aislados con las mayores actividades antifúngicas fueron también evaluados contra 2 cepas de Candida albicans resistentes a antifúngicos. Se realizó la secuenciación parcial del ARNr 16S y el análisis filogenético de los aislados que mostraron actividades antifúngicas de amplio espectro. Se seleccionaron 42 de 202 actinomicetos aislados por sus fuertes actividades antifúngicas. La mayor proporción de aislados de suelo rizosférico de plantas viejas mostraron antagonismo contra bacterias gram positivas y hongos fitopatógenos (proporciones de 0,259; 0,431 y 0,345 contra Fusarium oxysporum, Rhizoctonia solani y Pythium ultimum, respectivamente), mientras que la mayor actividad antagónica contra las bacterias gram negativas predominaron en aislados de suelo no asociado a raíces. Los aislados de suelo rizosférico de plantas jóvenes se caracterizaron por una fuerte actividad antagónica contra F. oxysporum (proporción de 0,333, p<0,05). El análisis filogenético de secuencias del ADNr 16S mostró que los aislados que presentaron fuerte actividad antifúngica fueron genéticamente similares. Concluimos que el suelo rizosférico de A.tridentata es una fuente excelente para el descubrimiento de actinomicetos productores de compuestos antifúngicos potencialmente novedosos.
Actinomycetes comprise an extensive and diverse group of gram-positive, aerobic, mycelial bacteria that play an important physiological and ecological role in soil.29,31 They can degrade a wide diversity of recalcitrant compounds such as lignocelluloses and many other polymers occurring in soil and litter, as well as a range of xenobiotic compounds.4,7,19 Because of their metabolic diversity, actinomycetes are a great source of lytic enzymes, antibiotics and a great deal of other bioactive metabolites.5,10,28 This group of organisms produce more than half of the naturally occurring antibiotics discovered to date and continue to be screened for useful compounds.5,8
Because actinomycetes are largely spread in nature, they have been isolated from many environments including soils, composts, plant materials and waters.5,10,23,26 Actinomycetes, especially Streptomyces spp. isolated from rhizosphere soils, sometimes represent novel species.16,20 They are great root colonizers, where they protect the plants against phytopathogens and promote plant growth.1,11,24,33
Desert plants such as Artemisia spp. are known to produce a great diversity of phenols and terpenoids with antimicrobial activities.30 Thus, rhizosphere actinomycetes from Artemisia, which can endure unfavorable growth conditions, are worthy of examination for antimicrobial activities.
The present study involved the screening for antimicrobial activities of actinomycetes from rhizosphere soils of Artemisia tridentata and their counterpart bulk soils. Their antagonistic activity was characterized based on in vitro bioassays against a broad panel of bacteria and fungal plant pathogens. A select group of the actinomycetes strongly antagonistic to all filamentous fungi tested was further characterized for antagonism toward two drug-resistant strains of Candida albicans: ATCC MY-204276 (fluconazole-resistant) and ATCC 44373 (5-fluorocytosine-resistant). In addition, a selection of three strongly antifungal groups of isolates were further characterized by partial 16s rRNA gene amplification, and phylogenetic trees were constructed to determine if there was a relationship between antimicrobial activity and genetic relatedness.
Material and methodsSource of actinomycetesTwo hundred and two actinomycete strains were previously isolated from two sagebrush rhizosphere soils [one from a young plant (RSYP) and one from an old growth plant (RSOP)], and two non-rhizosphere bulk soils near the sagebrush (B1Y and B1O, for the young and old plant system, respectively).9
Evaluation of antimicrobial activitiesAntimicrobial susceptibility tests were performed in vitro by using a panel of plant pathogen strains including: Fusarium oxysporum ATCC 070233 as well as Rhizoctonia solani and Pythium ultimum; two gram-positive bacteria (Bacillus subtilis and Rathayibacter tritici) and two gram-negative bacteria (Xanthomonas campestris pv. campestris and Burkholderia cepacea). All fungal strains were from Dr. Don Crawford's laboratory stock, Department of Microbiology, Molecular Biology and Biochemistry, while the bacterial strains were from the Bacteriology laboratory, Department of Entomology, Plant Pathology and Nematology.
The antimicrobial susceptibility for filamentous fungi were assessed following the in vitro plate bioassay. Actinomycete isolates were streak-inoculated to one side of PDA and YDA plates and incubated at 30°C for eight days to allow the production and diffusion of metabolites and extracellular hydrolytic enzymes. An agar plug containing actively growing fungus was then placed onto the opposite side of the inoculated plates and incubated at 30°C. Fungal mycelial plugs were placed on noninoculated plates as controls. Growth inhibition was recorded at different time intervals, depending on the fungus, for 7 days.
The antimicrobial bioassay plates for bacteria were performed by streak-inoculation of the actinomycete to one side of multiple PDA plates and incubated at 30°C for 10 days to allow the production and diffusion of metabolites and extracellular hydrolytic enzymes. Forty-eight-hour bacterial growth from NBY (ATCC Medium 763) plates was then inoculated as lines perpendicular to the actinomycete growth and incubated at 30°C. Bacteria were also streaked on non-inoculated plates to serve as controls.2 Bacterial growth inhibition was recorded at different time intervals for 5 days.
A selected group of actinomycetes strongly antagonistic to all filamentous fungi tested were further characterized for antagonism toward two drug-resistant strains of Candida albicans: ATCC MY-204276 (fluconazole resistant) and ATCC 44373 (5-fluorocytosine-resistant). Anti-Candida activity was tested as described for bacteria except for PDA and YDA were used instead.
PCR amplification of partial 16s rRNA gene sequencesGenomic DNA from the isolates with potent antifungal activity were extracted using the UltraClean™ Microbial DNA Isolation Kit (Mo Bio Laboratories, Inc., Solana Beach, CA) according to the manufacturer's instructions. 16S rDNA partial sequences of cultured isolates were amplified using the primers 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 907R (5ʹ-CCGTCAATTCMTTTRAGTT-3ʹ). Each PCR mixture contained 20μmol/l (each) primer, 0.2mmol/l (each) dNTPs, 25mmol/l MgCl2, 5μl of 10x PCR universal buffer (Invitrogen TECH-LINE™, USA) 1μl of the DNA template, and 1.25U of Taq DNA polymerase (Invitrogen TECH-LINE™, USA) to a final volume of 50μl.
Thermocycling conditions were as follows: one cycle at 95°C for 5min; followed by 30 cycles (each) at 95°C for 1min, 55°C for 1min, and 72°C for 2min; and finally, one cycle at 72°C for 7min in a Gene Amp PCR System 2400 thermocycler (Applied Biosystems). The positive control consisted of reaction mixtures containing 8μg of DNA of Streptomyces lydicus WYEC-108. The negative control lacked the DNA template but contained all other reactants. A 1-kb plus ladder (GIBCO BRL Life Technologies) was used as a DNA size marker.
The purified PCR amplicons were sequenced by the Laboratory of Biotechnology and Bioanalysis at Washington State University (Pullman, WA). Sequences were compared against known sequences using the NCBI BLAST database.
Phylogenetic analysisThe BioEdit program was used as an editing tool to facilitate sequence analysis. Multiple alignments were obtained using the Clustal W program. Phylogenetic trees were inferred by three algorithms, the maximum-parsimony, neighbor-joining, and maximum-likelihood methods using the PAUP* package. In addition, the 16S rDNA sequence of Haemophilus paragallinarum was added to the analysis as an out-group. Bootstrap analyses for the neighbor-joining and the maximum-likelihood results were generated based on 200 re-samplings.
Evolutionary distance matrices for the neighbor-joining method were generated. The TreeView (WIN 32) program was used for viewing the trees generated by the three algorithms.
Statistical analysisThe generalized linear model was used, assuming a binomial distribution to test for significant activity effect across soils. Pair-wise comparisons among soils were done using contrasts and chi-square tests. All computations were carried out using SAS 8.2 Copyright (c) 1999–2001 by SAS Institute Inc., Cary, NC, USA.
ResultsTwo hundred and two actinomycetes were isolated from the four mentioned soils and their antifungal activities showed that isolates from both rhizosphere soils, RSYP and RSOP, had the highest activity against F. oxysporum; however, they were not significantly different from those of their bulk soils (Table 1). The highest antagonistic activities against R. solani were detected in RSOP; this was significantly different from the rest of the soils, followed by the activities of bulk soils B1Y and B1O. The highest anti-Pythium activities were detected in organisms found in three soil samples: bulk soil B1Y and rhizosphere soils, RSOP and RSYP. Bulk soil B1O activity was significantly lower than that in the other three soils (Table 1).The antibacterial activities of isolates from bulk soils B1Y and B1O had higher activities against Gram-negative bacteria, X. campestris pv. campestris and B. cepacea than their counterparts RSYP and RSOP; however, no statistical differences were detected among soils (Table 2). On the other hand, the highest activities against Gram-positive bacteria, such as B. subtilis, were detected in the isolates of bulk soil B1Y and rhizosphere soil RSOP, with no statistical difference between the two, followed by the antagonistic activity of bulk soil B1O (Table 3). Finally, isolates from soil B1Y showed the highest antagonistic activity against Rathayibacter, followed by both rhizosphere soils, RSYP and RSOP; isolates from bulk soil B1O had the lowest activity against Rathayibacter (Table 2).
Distribution of antifungal activities in four soils of a sagebrush habitat.
| Soil samples | Active isolates against | ||
|---|---|---|---|
| F. oxysporum | R. solani | P. ultimum | |
| RSYP | 0.333a | 0.074b | 0.333a |
| B1Y | 0.203a | 0.257b | 0.487a |
| RSOP | 0.259a | 0.431a | 0.345a |
| B1O | 0.209a | 0.256b | 0.140b |
The data presented are proportions of the active isolates with respect to the total population in each soil (RSYP, n=27; B1Y, n=74; RSOP, n=58; B1O, n=43). RSYP and RSOP are rhizosphere soils from a young and old sagebrush plants, respectively; B1Y and B1O are their counterpart bulk soils, respectively. Different letters within a column indicate significant difference at p<0.05. All comparisons are based on logit transformations [(P/1−P), where P is the transformation values of active isolates].
Distribution of antibacterial activities in four soils of a sagebrush habitat.
| Soil samples | Active isolates against | |||
|---|---|---|---|---|
| B. subtilis | R. tritici | X. campestris pv. campestris | Burkholderia cepacea | |
| RSYP | 0.185c | 0.260b | 0.222 a | 0.037a |
| B1Y | 0.527a | 0.500a | 0.338 a | 0.243a |
| RSOP | 0.483a | 0.224b | 0.103a | 0.052a |
| B1O | 0.209b | 0.023c | 0.163a | 0.116a |
The data presented are proportions of the active isolates with respect to the total population in each soil (RSYP, n=27; B1Y, n=74; RSOP, n=58; B1O, n=43). RSYP and RSOP are rhizosphere soils from a young and an old sagebrush plants respectively; B1Y and B1O are the counterpart bulk soils respectively. Different letters within a column indicate a significant difference at p<0.05. All comparisons are based on logit transformations.
Blast search of partial sequences of the 16s RNA gene of selected isolates.
| Isolatesa | Match | Identityb (%) | GenBank accession number |
|---|---|---|---|
| Group 1 | |||
| R1Y9 | Streptomyces kasugaensis | 721/725 (99%) | DQ629032 |
| R1Y10 | Streptomyces kasugaensis | 786/791 (99%) | DQ629033 |
| B1Y14 | Streptomyces cf. griseusStreptomyces argenteolus | 645/648 (99%) | DQ629050 |
| B1Y54 | Streptomyces violaceusniger | 756/764 (98%) | DQ629051 |
| B1Y71 | Streptomyces sp. IM-8062 | 710/715 (99%) | DQ629052 |
| R1O41 | Streptomyces sp. KACC 91020 | 776/791 (98%) | DQ629031 |
| R1O44 | Actinobacterium 17a-5 | 684/688 (99%) | DQ642601 |
| R1O3-2 | Streptomyces sp. LK4-2 | 706/713 (99%) | DQ629028 |
| R1O4-2 | Streptomyces sp. LK4-2 | 680/687 (98%) | DQ629029 |
| R1O7-2 | Streptomyces sp. LK4-2 | 709/716 (99%) | DQ629030 |
| B1O9 | Streptomyces erumpens | 771/773(99%) | DQ629048 |
| B1O10 | Streptomyces luteogriseusActinobacterium 17a-5 | 768/772 (99%)765/768 (99%) | DQ629049 |
| Group 2 | |||
| R1Y11 | Actinobacterium 17a-5 | 740/742 (99%) | DQ629038 |
| R1Y25 | Streptomyces griseocarneus | 771/782 (98%) | DQ629039 |
| B1Y4 | Actinobacterium 17a-5 | 767/771 (99%) | DQ629053 |
| B1Y37 | Streptomyces ciscaucasicus | 732/732 (100%) | DQ629054 |
| B1Y40 | Actinobacterium 17a-5 | 740/742 (99%) | DQ629055 |
| B1Y42 | Stretomyces sp. KN-0647 | 724/730 (99%) | DQ629056 |
| B1Y43 | Stretomyces sp. KN-0647 | 726/732 (99%) | DQ629057 |
| R1O4 | Actinobacterium 17a-5 | 767/770 (99%) | DQ629034 |
| R1O23 | Actinobacterium 17a-5 | 737/739 (99%) | DQ629035 |
| R1O24 | Streptomyces cyaneus | 703/704 (99%) | DQ629036 |
| R1O46 | Streptomyces turgidiscabies | 681/686 (99%) | DQ629037 |
| Group 3 | |||
| B1Y2 | Streptomyces sp. IM-8062 | 763/769 (99%) | DQ629062 |
| B1Y3 | Actinobacterium 17a-5 | 733/733 (100%) | DQ629063 |
| B1Y16 | Streptomyces sp. KN-0647 | 730/736 (99%) | DQ629064 |
| B1Y21 | Streptomyces sp. IM-6899 | 729/740 (98%) | DQ629065 |
| B1Y33 | Streptomyces sp. KN-0647 | 746/752 (99%) | DQ629066 |
| B1Y48 | Streptomyces sp. IM-8062 | 683/688 (99%) | DQ629067 |
| B1Y64 | Streptomyces platensisS. catenulaeS. tubercidicus | 765/778 (98%)765/778 (98%)765/778 (98%) | DQ629068 |
| R1O3 | Streptomyces sp. Sm22Streptomyces sp. IM-6784 | 657/661 (99%) | DQ629040 |
| R1O31 | Streptomyces sp. | 709/718 (98%) | DQ629041 |
| R1O195 | Actinobacterium 17a-5 | 737/739 (99%) | DQ629047 |
| R1O37 | Streptomyces lincolnensisStreptomyces ciscaucasicus | 721/723 (99%) | DQ629042 |
| R1O45 | S. peruviensisS. ciscaucasicus | 706/713 (99%) | DQ629043 |
| R1O49 | Streptomyces africanus | 655/655 (100%) | DQ629044 |
| R1O54 | Actinobacterium 17a-5 | 661/661 (100%) | DQ629045 |
| R1O59 | Actinobacterium 17a-5 | 722/722 (100%) | DQ629046 |
| B1O22 | Streptomyces coerulescensStreptomyces bellus | 749/750 (99%) | DQ629058 |
| B1O26 | Streptomyces sp. | 745/750 (99%) | DQ629059 |
| B1O27 | Streptomyces sp. | 693/697 (99%) | DQ629060 |
| B1O32 | Actinobacterium 17a-5 | 734/741 (99%) | DQ629061 |
The isolates were the strongest fungal inhibitors from 202 isolates of a sagebrush habitat. They were divided into three groups: antagonistic isolates toward Pythium, Rhizoctonia and Fusarium (Group 1), isolates with anti-Pythium activities (Group 2), and those with anti-Rhizoctonia and anti-Fusarium activities (Group 3).
Forty-two isolates out of two hundred and two actinomycetes were selected for their strong antifungal activity. They were divided into three groups: Group 1 was comprised of antagonistic isolates with broad-spectrum antifungal activity against lower (P. ultimum) and higher filamentous fungi (F. oxysporum and R. solani); Group 2 contained isolates with only anti-Pythium activity, and Group 3 was comprised of those isolates with both anti-Rhizoctonia and anti-Fusarium activities (Table 3). Of the selected strains, 17 were from RSOP (overall 29.3% of isolates with strong antifungal activity); bulk soil B1O accounted for six isolates (overall 13.95% of isolates with strong antifungal activity); none of the isolates of this soil showed strong inhibition against P. ultimum. On the other hand, only four isolates (overall 14.8% of isolates with strong antifungal activity) from RSYP were selected; none of the isolates were classified into Group 3. Its counterpart bulk soil BYO accounted for 15 strong antifungal isolates (20.3% of isolates with strong antifungal activity).
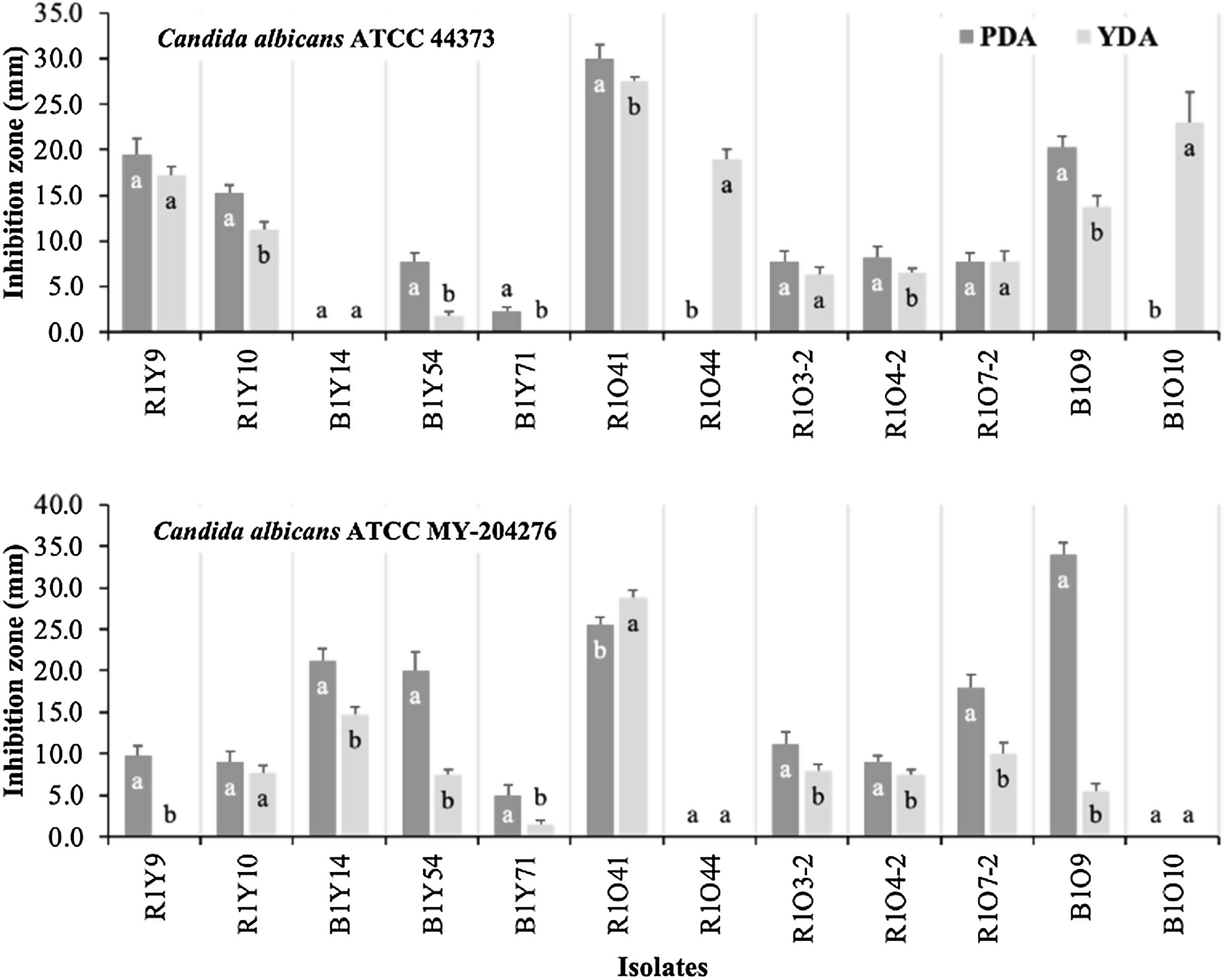
Anti-Candida activitiesTwelve isolates that showed antagonism against all three phytopathogenic filamentous fungi tested were also screened for anti-Candida activities (Fig. 1). Antagonistic activity was detected toward both strains of C. albicans (ATCC MY-204276, fluconazole-resistant and ATCC 44373, 5-fluorocytosine-resistant) in both media (PDA and YDA), with different levels of activities observed between both media. C. albicans ATCC 44373 was inhibited to the greatest extent on PDA by strain R1O41, followed by B1O9, R1Y9 and R1Y10 while isolates R1O44 and B1O10 only showed antagonistic activity on YDA, but to a great extent, after R1O41. Isolates R1O7-2, R1O3-2, R1O4-2 and B1Y54, all showed lesser antagonistic activity in both media (Fig. 1A). Inhibition toward C. albicans ATCC MY-204276 was higher on PDA in most isolates (Fig. 1). On PDA, strain R1O41 was the best inhibitor, followed by B1Y14, B1Y54 and R1O7-2 while isolates R1O41, B1Y14 and R1O7-2 showed the highest activity on YDA; isolate R1Y9 only showed antagonism on PDA. The comparison of anti-Candida activity on PDA and YDA by strain is shown in Figure 1.
Anti-Candida activity of selected actinomycetes from four soils of a sagebrush habitat in two different media. The data presented are the means of quadruplicate measurements of inhibition zones. Bars represent their standard deviations. Mean values with equal letters are not statistically different (Tukey, p<0.05) within each isolate.
Analysis of the partial 16S rRNA gene sequences of 42 selected strong antifungal isolates by BLAST revealed that 31 belonged to the genus Streptomyces while the remaining 11 strains were only identified as actinobacteria (Table 3). Analysis of the 16S rRNA gene of the isolates B1Y37 and R1O49 showed 100% sequence identity with Streptomyces ciscaucasicus strain DSM 40275 (accession AY508512), and Streptomycesafricanus (accession AY208912), respectively. Isolates B1Y3, R1054 and R1O59 had 100% sequence identity with the undefined actinobacterium 17a-5 (accession AY561563). Isolates B1Y54, R2O4, R1Y25, B1Y64, and R1O31 had 98% sequence identity to S. violaceusniger (accession AJ391823), Streptomyces sp. LK4-2 (AY277376), S. griseocarneus (X99943), Streptomyces sp. (accession AY167807, AJ621613 and AJ621604), and Streptomyces sp. (Y15499), respectively. The remaining isolates had 99% sequence identity with the best match(es) in the blast search (Table 3).
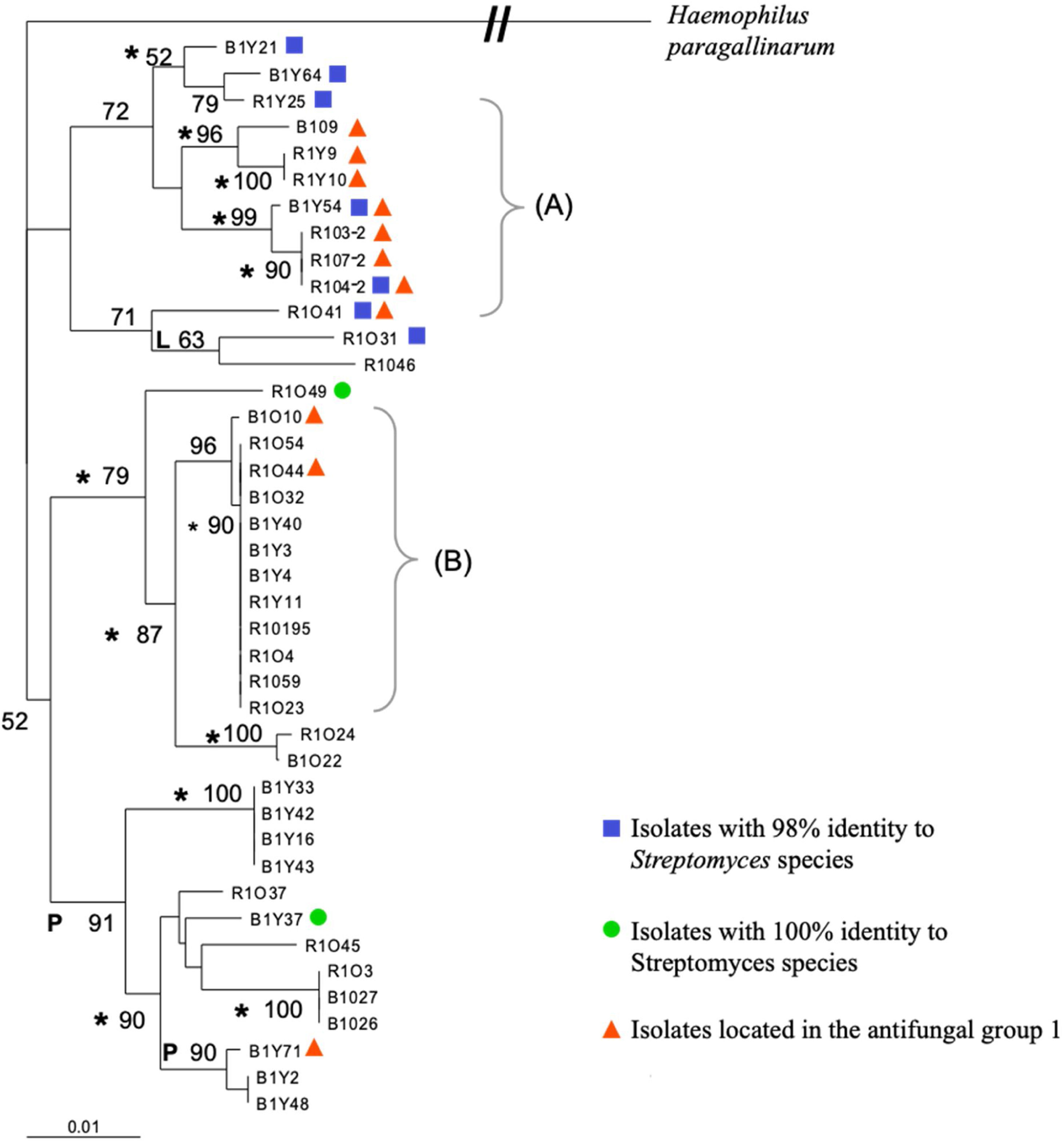
Fragment 27F-907R of the forty-two selected isolates was also used to construct a multiple alignment and a phylogenetic tree, which is shown in Figure 2. Bootstrap analysis of the tree revealed that all of the phylogenetic relationships were not resolved by using only partial 16S rDNA sequences. Additional studies will be needed to resolve all the intraspecies relationships among various Streptomyces strains; however, very defined branches and some clades were generated with the three different algorithms used. One of the defined branches grouped all the isolates identified in the blast search as actinobacteria, while those isolates with 98% identity to Streptomyces sp. were in one of the major clusters of the neighbor-joining tree (Fig. 2). All the antagonists against filamentous fungi of group one that also showed anti-yeast activity were located on one major cluster by the analysis. Those isolates within group two (strong anti-Pythium activity) and group three (strong anti-Rhizoctonia and Anti-Fusarium activity), were found all over the tree, and did not fall into a specific branch or clade on the tree (Fig. 2).
Phylogenetic relationship of partial 16S rRNA gene sequences of 41 streptomycetes in a neighbor-joining tree. L and P indicate branches that were also found when we used the maximum-likelihood and maximum-parsimony methods, respectively; the asterisks indicate branches recovered with all three methods. The numbers at the nodes indicate the level of bootstrap support based on a neighbor-joining analysis; only values that were >50% are given. The scale bar indicates 0.01 substitutions per nucleotide position. The bracket (A) indicates the isolates with antimicrobial activity against filamentous fungi and C. albicans and (B) indicates the clade of isolates identified with actinobacterium 17a-5.
Within the rhizosphere, plant roots have a direct effect on composition and density of the soil microbial populations. Root exudates selectively influence the growth of bacterial and fungal populations by altering the presence of substrates in soil in the vicinity of roots.27 The varieties of organic compounds released by plants have been postulated to be a key factor influencing the diversity of microorganisms in the rhizosphere of different plant species.3,32 Sagebrush roots are known for their wide production of phenols, and other aromatic compounds as well as terpenoids with antimicrobial activities.13,17,30 Those antimicrobial compounds can be used as a carbon source by some microorganisms, including actinomycetes.12,19 Therefore, it is not surprising that actinomycetes actively grow and colonize root systems like those of desert sagebrush plants.9,15
In the present work, the sagebrush rhizosphere soil of the old plant (RSOP) appears to be enriched in highly active antifungal and antibacterial Gram-positive compound-producing actinomycetes; while the rhizosphere soil of the young plant (RSYP) enriched for anti-Fusarium actinomycetes with lower antibacterial actinomycetes compared to its counterpart bulk soil showing a lower rhizosphere effect than that in the old plant. These differences can be explained by the rhizosphere effect, and by the qualitative and quantitative differences in root exudates due to the different plant ages.21,27 In this context, it was demonstrated that the roots of Artemisia tridentata produce different types of antimicrobial and phytotoxic secondary metabolites depending on the age of the plant.13 On the other hand, in our study, the antimicrobial activities of each rhizosphere soil were compared to its counterpart bulk soil to avoid soil effect. It is well known that the soil type qualitatively and quantitatively affects microbial communities in the rhizosphere.3
Forty-two actinomycetes (21% of the total number of isolates) were selected for their strong antifungal properties and classified into three groups based on their activities toward F. oxysporium and R. solani (higher fungi rich in chitin in their cell wall), and P. ultimum (a lower fungus rich in cellulose in its cell wall).
Isolates from the four soils were present in each of the three antifungal groups, except for isolates from RSYP and B1O. RSYP isolates showed strong antagonism against R. solani or F. oxysporum, but none of them showed antagonism against both fungi while B1O isolates did not show strong antagonism, only against P. ultimum.
In our study of anti-yeast activity, diverse results were observed. Isolate R1O41 showed the strongest antagonism in both media compared to the remaining antagonistic isolates evidencing the potent broad-spectrum antifungal activities and their complex strategies to control the two antifungal drug-resistant Candida albicans strains. A significant difference (p<0.05) in anti-yeast activities between media within the same antagonist was observed in some of them, which may be influenced by diverse factors that involved the synthesis of antibiotics and other secondary metabolites in culture media. Data previously published showed that medium composition and their concentrations are strongly related to antibiotic production.22 Simply metabolizable carbon sources such as glucose generally repress production of many antibiotics, particularly when they are used as the sole carbon source.25 Studies on fermentation media show that polysaccharides are generally the best carbon sources for antibiotic production as they support a slow growth rate which is desirable for antibiotic production.6,22,25 Moreover, there are also cases where glucose is an excellent carbon source for antibiotic production. It has been reported that dextrose was a great carbon source for the antibiotic production of Streptomyces kanamyceticus M27.18
Nitrogen is another component strongly related to antibiotic synthesis. As with the carbon component, simply metabolizable nitrogen sources usually decrease antibiotic production while complex nitrogen sources such as yeast extract, malt extract and soybean meal can increase the production of antibiotics produced by streptomycetes, which can be attributed to the slow decomposition of these compounds in the medium.18,22
In addition to nutrients, microbial interactions can regulate the production of antibiotics and other secondary metabolites. In such interactions, production of secondary metabolites can facilitate communication, but can also act as defensive molecules which help microorganisms to defend themselves against competitors.31 In our study of the anti-yeast bioassay, the signaling between the yeast and each antagonist must be unique in each case and may be reflected in some antifungal activities or the lack of them; for example, B1Y14 showed antifungal activity against C. albicans ATCC MY-204276 in both media; however, no activity was detected against C. albicans ATCC 44373. Similarly, other isolates, R1O44 and B1010, showed activity against C. albicans ATCC 44373 in YDA; however, no activity was observed against C. albicans ATCC MY-204276.
In some studies, mainly members of the genus Streptomyces were detected by screening for antifungal actinomycetes from the rhizosphere soil of different plants including medicinal and forage plants.14,33 Similarly, our results of the 16S rDNA analyses revealed that most of the selected antifungal isolates belong to the genus Streptomyces, except for eleven isolates that showed a best identity match with actinobacterium 17a-5. After actinobacterium 17a-5, Streptomyces luteogriseus and S. tuirus followed in the search, each with lower scores and percent sequence identity (data not shown), suggesting that those eleven isolates may be members of a new Streptomyces species not yet identified. Furthermore, the phylogenetic analysis showed that all the isolates identified with actinobacterium 17a-5 were in a very defined terminal clade. On the other hand, all the antifungal isolates with 98% identity with Streptomyces sp. were in one of the major clusters of the neighbor-joining tree; they are potentially novel species; however, further analysis are required to determine that. In addition, almost all the strains of antifungal group one, which also exhibited anti-Candida activity (Fig. 1), were in close proximity on the phylogenetic tree (R1O41, R1O4-2, R1O7-2, R1O4-3-2 B1Y54, R1Y10, R1Y9 and B1O9), suggesting the presence of a potential clade of strong antifungal Streptomyces with potential novel bioactive metabolites. The isolate denominated R1O41, isolated from RSOP, seems to be a novel actinomycete isolate with strong broad antifungal activity against the three phytopathogens and the two drug-resistant Candida strains, showing a great potential that can be exploited for use in agriculture and medicine.
ConclusionsOur study showed that the rhizosphere soil of sagebrush plants may preferentially favor antifungal actinomycete colonizers, particularly plants with high rhizosphere effect. The isolates with strong antifungal activities were identified as members of the genus Streptomyces. A relationship was detected by phylogenetic analysis between the genetic relatedness and the antifungal activities of those actinomycetes exhibiting strong antifungal activity against all the filamentous fungi tested along with anti-Candida activity. Finally, the rhizosphere soil of A. tridentata seems to be a great source of novel actinomycetes with strong antifungal activity that can be exploited for use in agriculture and medicine.
Conflict of interestThe authors declare that they have no conflicts of interest.
The authors sincerely thank Dr. Don Crawford, College of Agriculture, University of Idaho, for technical advice in this research. Also, the authors thank Dr. Celeste Brown for helping in the phylogenetic analysis. This research was supported in part by the “Programa para el Desarrollo Profesional Docente, para el Tipo Superior (PRODEP)” of Mexico, grant UACHIH-99-05-01.