Increasing antimicrobial resistance among Staphylococcus aureus necessitates a new antimicrobial with a different site of action. We have isolated a novel cyclic peptide-1 (ASP-1) from Bacillussubtilis with potent activity against methicillin-resistant S. aureus (MRSA) at a minimum inhibitory concentration (MIC) of 8–64μg/ml. Scanning electron micrographs demonstrated drastic changes in the cellular architecture of ASP-1 treated cells of S. aureus ATCC 29213 and an MRSA clinical isolate at MICs, with damages to the cell wall, membrane lysis and probable leakage of cytoplasmic contents at minimum bactericidal concentrations. The ultrastructure alterations induced by ASP-1 have also been compared with those of oxacillin-treated MRSA cells at its MIC using scanning electron microscopy.
El incremento de la resistencia antimicrobiana entre los tipos de S. aureus exige un nuevo agente antimicrobiano con un sitio de acción diferente. Aislamos un nuevo péptido cíclico (ASP-1) de Bacillussubtilis con potente actividad frente a S. aureus resistente a meticilina (SARM) en una concentración inhibitoria mínima (CIM) de 8-64μg/ml. Las micrografías obtenidas con microscopio electrónico de barrido mostraron cambios drásticos en la arquitectura celular de las células de S. aureus ATCC 29213 tratadas con ASP-1, y un aislamiento clínico de SARM a la CIM, con daños a la pared celular, lisis de la membrana y probable fuga de contenido citoplasmático a concentraciones bactericidas mínimas. Comparamos también, las alteraciones de la ultraestructura inducidas por ASP-1 con las de células de SARM tratadas con oxacilina a su CIM, utilizando microscopio electrónico de barrido.
Emerging and re-emerging “superbug” infections are a global threat due to indiscriminate use of antibiotics. Methicillin-resistant Staphylococcus aureus (MRSA), one of the most lethal causative agents, has spread rapidly all over the world. To combat this global menace, the search for alternative antimicrobials is a pressing need, especially since most antimicrobials (glycopeptides) have exhibited decreased efficacy and susceptibilities against multidrug-resistant gram-positive infections. Antimicrobial peptides (AMPs) are gaining attention as a potential alternative to conventional antibiotics and small molecule drugs6–8. Earlier studies indicated that the cyclic molecules are among the most effective antimicrobials6,11. These studies have rendered AMPs as promising new classes of antibiotic candidates. A critical analysis of the altered ultrastructure of the cell membrane or the cell wall by electron microscopy (EM) enables us to understand the mechanisms of cell damage at sub-lethal to lethal concentrations of antimicrobial peptides. Earlier, we have reported the antibacterial spectrum and potency of ASP-1 against 29 bacterial isolates, including 12 MRSA isolates2,3. Our recent study revealed the narrow-spectrum of cyclic ASP-1 against methicillin/oxacillin resistant Staphylococcus (ORSA) and biofilm-forming bacteria had a lower chance of developing resistance3. The present study was aimed at determining the effect of reversed-phase high-performance liquid chromatography (RP-HPLC)-purified ASP-1 on the ultrastructure of MRSA-4, a clinical isolate along with a reference strain S. aureus ATCC 29213 by scanning electron microscopy (SEM) and mode of action using a time-kill assay against a selected MRSA isolate.
The purification of ASP-1 was determined by a multistep process involving acid precipitation, solvent extraction, and adsorption with silica gel chromatography (230–400 mesh size) followed by semi-preparative and analytical reversed-phase high-performance liquid chromatography (RP-HPLC) as described earlier2,3. The purification process started with the preparation of cell-free supernatant (CFS) from the freshly cultured cells of the producer strain Bacillussubtilis URID 12.1, which were grown at 37°C for 44h at 110rpm in modified tryptone soy broth (mTSB, pH 7.4±0.2), and the CFS was prepared by centrifugation at 12000rpm, 4°C for 20min. The bioactive fractions were pooled and further purified by RP-HPLC using a C18 column (4.6mm×250mm, particle size 5μm, Agilent, Santa Clara, CA, USA). The eluted active fraction with a retention time of 30.5min (data not shown) indicates its non-polar nature. The peptide purified at a semi-preparative scale was collected and tested for antistaphylococcal activity. These pooled active fractions, when subjected to re-chromatography on an analytical column, showed a single peak indicative of the pure fraction obtained.
In our previous study, we reported the antibacterial activity of ASP-1 against several MRSA isolates at a MIC of 0.3–16μg/ml and MBC of 2–128μg/ml, to be better than oxacillin2,3. In the present study, a total of 12 MRSA clinical isolates were used along with a reference strain S. aureus ATCC 29213 to confirm the presence of two antibiotic resistance genes in MRSA isolates and one housekeeping gene specific to S. aureus. The bacterial cells were grown in Müeller Hinton Broth (MHB)/Agar (MHA, HiMedia, Mumbai, India). DNA was extracted from the standard test strain and test isolates using Power Lyzer® DNA isolation kit (MOBIO, HiMedia, Mumbai, India) following the manufacturer's protocol. Nucleic acid concentrations were estimated by NanoDrop™ spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). All indicator strains used in our previous study2,3 were subjected to the detection of mecA, femA and nuc genes by real-time (rt)-PCR as described by Li et al.13 using the following primers: mecA1-5′-AAAATCGATGGTAAAGGTTGGC-3′, mecA2-5′-AGTTCTGCAGTACCGGATTTGC-3′14; nuc1-5′-GCGATTGATGGTGATACGGTT-3′, nuc2-5′-AGCCAAGCCTTGACGAACTAAAGC-3′10, and femA1-5′-CAGTCACTAGCTGGGCCACTT-3′ and femA2-5′-GTCAATTAGGTTATGTCGGAGGA-3′1. The amplification reaction was performed with Fast Start Universal SYBR Green Master (Roche Diagnostics, Indianapolis, IN, US) using 1μl of template and 40pmol each of forward and reverse primers. The gene targets were subjected to thermal cycling as follows: for 10min at 94°C and then 40 cycles of 30s at 94°C, 30s at 55°C, and 60s at 72°C for the mecA gene primer set; 30 cycles of 45s at 94°C and 45s at 54°C, and 30s at 72°C for femA gene primer set; 30 cycles of 30s at 94°C, 30s at 55°C, and 60s at 72°C for nuc gene primer set. The reaction was performed by using a LightCycler® 96 Real-Time PCR system (Roche Diagnostics). The PCR products were then confirmed by 2% agarose gel electrophoresis.
To determine the in vitro antimicrobial activity of the test agent over time, i.e., the pharmacodynamics of the test agent (killing rate at different concentrations and time; or concentration- and time-dependent bactericidal or bacteriostatic activities), time-kill assays were conducted. MRSA-153 (renamed as MRSA-5 in the present study) cells were subcultured in cation-adjusted Mueller–Hinton broth (CAMHB) (Hi-Media, India). Inoculum preparation, inoculation, and determination of the microbial count at respective MIC and MBC were performed as described previously3,15. MRSA-15 bacterial cells were washed using saline and adjusted to ∼5×104CFU/ml in CA-MHB and the test agent ASP-1 was added at final concentrations of 1× and 5× MIC and incubated at 37°C. In order to study the morphological and ultrastructural changes, S. aureus ATCC 29213 and MRSA-4 cells were treated with ASP-1 at different concentrations and the changes were compared with the untreated cells of the two strains respectively using SEM. Bacterial cultures (108CFU/ml) were treated with ASP-1 at 1× MIC and 1× MBC for 0–12h and supernatant was drawn at hourly intervals. ASP-1-treated cells were then fixed on a clean grease-free coverslip using 2.5% glutaraldehyde in sterile phosphate-buffered saline (PBS, pH 7.3) at 4°C for 16h. Samples were washed thrice with sterile distilled water for 10min each, followed by treatment with osmium tetroxide as described elsewhere10. Subsequently, the samples were washed thrice with distilled water for 10min each; then treated with acetone gradient (30% to 100%) followed by critical point drying using CPD300 auto-program (Leica EM CPD300, Wetzlar, Germany). The dried specimens were mounted on stubs containing carbon adhesives and sputter-coated (Leica EM ACE600, Wetzlar, Germany) with gold. The images were captured by a Quanta 250 FEG (Thermo Fisher Scientific) at the SEM Facility, BITS Pilani, K. K. Birla Goa Campus, India.
Using the rt-PCR based identification approach, the presence of nuc and femA (Supplementary Information Fig. S1A), and mecA, nuc and femA genes (Supplementary Information Fig. S1B–D) was confirmed in all the 12 test isolates found to be resistant to oxacillin3. The amplification curves (Supplementary Information Fig. S1B–D) confirmed unequivocally the presence of the mecA, nuc and femA genes in all the MRSA isolates. The presence of the 300bp amplicon of the gene femA, and 270bp amplicon of the housekeeping nuc gene determined by agarose gel electrophoresis confirmed the identity of all the test isolates and reference strain as being S. aureus (Supplementary Information Fig. S2A and 2B). The presence of the femA gene in S. aureus ATCC29213 indicated that the femA gene could also be present in methicillin-sensitive (mecA-negative) S.aureus16. The mecA gene encodes for penicillin-binding protein-2 (PBP-2), and the femA gene is responsible for penta-glycine inter-peptide bridge formation that is essential for cell wall peptidoglycan synthesis; these two activities together are responsible for a higher level of methicillin-resistance in mecA-positive S.aureus13,16.
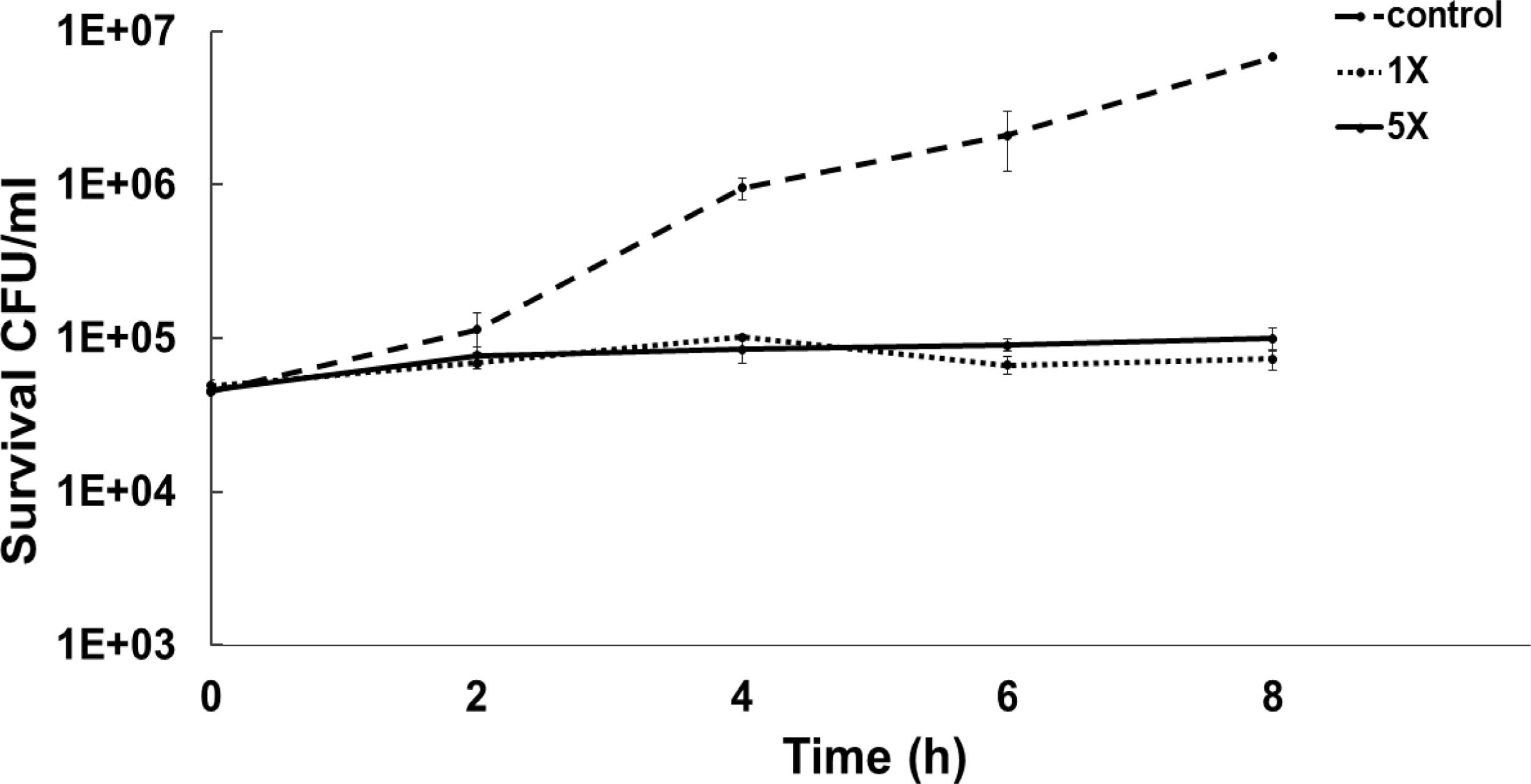
To determine the bacteriostatic or bactericidal nature of ASP-1 by a time-kill study we used MRSA-5 due to its sensitivity (MIC=16μg/ml) to test agent ASP-1 and resistance to oxacillin (MIC=1024μg/ml) revealed by the previous study3. The results in the form of a time-kill curve showed that our test agent ASP-1 was bacteriostatic at 1–5×MIC (Fig. 1), indicating that ASP-1 can possibly be used in combination with existing antibiotics for the treatment of highly resistant MRSA infections.
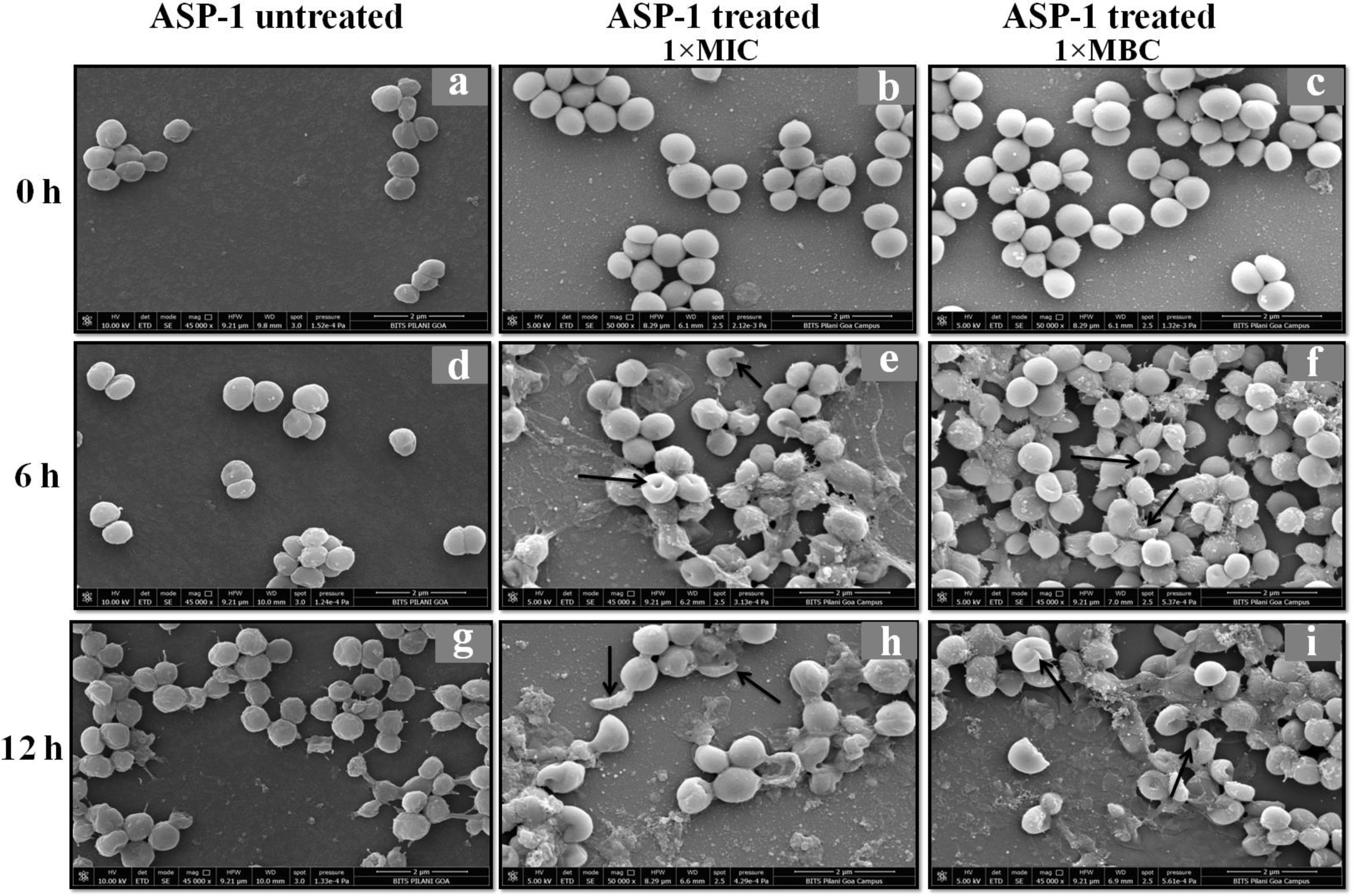
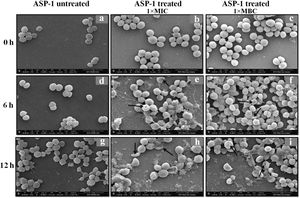
Previously, the time-kill curve of ASP-1 treated S. aureus ATCC 29213 cells showed reductions of 98.68% and 99.02% at 1× and 5× MICs respectively after 12h incubation3. Accordingly, the SEM images of ASP-treated ATCC29213 cells after 6 and 12h at 1× MIC and MBC (16μg/ml) showed extensive damages to the cell surface and distorted cell morphology, bringing forth interesting ultrastructure alterations (Fig. 2e, f, h and i). However, ASP-1 treated bacterial cells at 0h showed smooth cell surfaces with no discernible ultrastructural changes (Fig. 2b and c) and no ultrastructure alterations were observed in ASP-1 untreated cells at the three different time points (Fig. 2a, d and g).
SEM images of ASP-1 (1× MIC and 1× MBC) treated and untreated S. aureus ATCC29213 cells. Normal morphology of ASP-1 untreated S. aureus ATCC29213 cells (45000×) at different time points (a, d, and g); ASP-1 treated bacterial cells at 0h showing smooth cell surfaces with no discernible ultrastructural changes (b and c); ASP-1 treated cells showing deformities and central perforations within 6h of exposure (e and f); the SEM image reveals extensive cell surface damage at 12h as indicated by spurting of cellular contents and loss of shape (bending) with the central hollowing (h); several biconcave cells with central depression and cavity showing concomitant leakage of cytoplasmic contents at 1× MBC (16μg/ml); the field also shows some distorted cells lying at the periphery (i), and cells with dents, surface blebs and plicated surfaces have been observed. Several cells appeared as punctured with spewed-off cytoplasmic contents. Black solid arrows have indicated major structural changes.
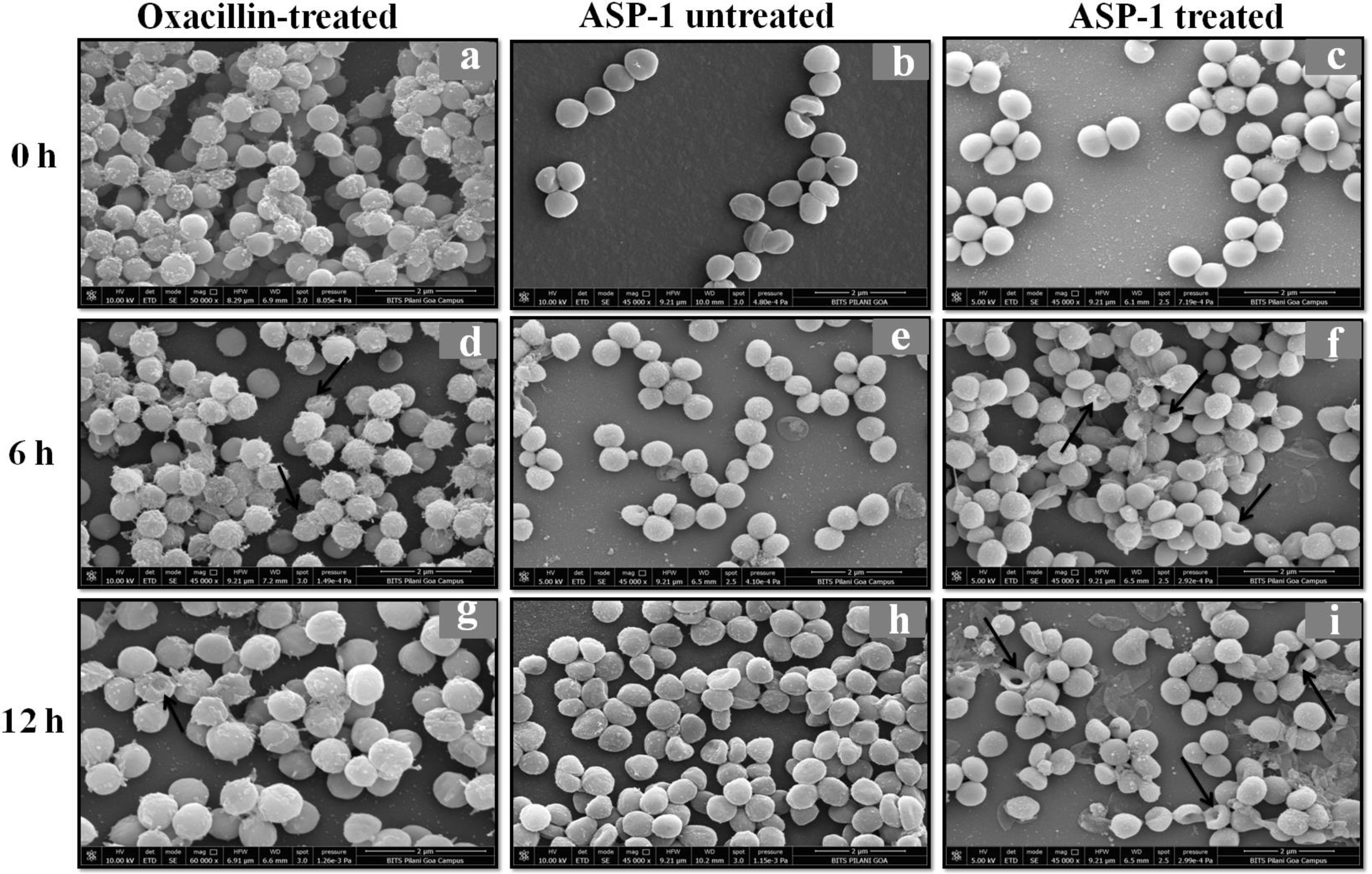
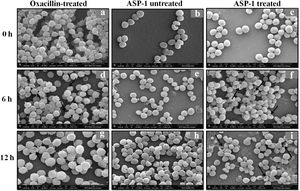
For the present SEM study, the MRSA-4 isolate was selected for the ASP-1 treatment as this clinical isolate, which is resistant to oxacillin (32μg/ml), cefoxitin (>8μg/ml), vancomycin (>16μg/ml), mupirocin (>4μg/ml) and amoxicillin/clavulanate (>4/2μg/ml), was found to be susceptible to ASP-1 (MIC=8μg/ml)2,3. ASP-1 untreated bacterial cells were healthy with nearly spherical shape and smooth intact surface, and in the actively dividing stage showing the presence of septa at different time points (Fig. 3b, e and h). Scanning electron micrographs of ASP-1 treated cells (Fig. 3f and i) revealed surface deformities though cells at 0h showed normal morphology (Fig. 3c). In some fields, several cells showed minor depression on the cell surface, which might be due to vacuum artifacts during SEM processing; a similar observation was also previously noticed in S. aureus cells after SEM processing9.
SEM images of oxacillin-treated, ASP-1 (1× MIC and 1× MBC) treated and untreated MRSA-4 cells. The SEM images of oxacillin (1× MIC=32μg/ml)-treated MRSA-4 cells captured at 0, 6 and 12h (a, d, and g); though few cells appear healthy as revealed by septum formation, other cells turned biconcave and deformed, and a single cell lying near the center showing a circular depression at 12h (d and g); SEM images of untreated MRSA-4 cells showing normal morphology and several healthy cells at dividing stage (b, e, and h); micrographs of ASP-1 treated cells (c, f, and i); cells showing normal morphology at 0h (c); biconcave and elliptical cells exhibiting central collapses, deep craters and deformations (f–i); and ASP-1 (1× MBC=16μg/ml) treated bacterial cells at 12h, showing centrally collapsed biconcave cells with spewed-off contents after cell lysis (i). Black solid arrows have indicated major structural changes.
The scanning electron micrographs (Fig. 2e and h) of ASP-1 (1×MIC=8μg/ml, 1×MBC=16μg/ml)-treated bacterial cells showed membrane distortions with surface depressions, and biconcave appearances with dents compared to ASP-1 untreated cells (Fig. 2a, d and g) at three different time points 0, 6 and 12h. The formation of holes on the cell surface and leakages of cellular contents were visible when the cells were incubated for 12h with ASP-1 at the same concentration (Fig. 2h). A similar ultrastructural change was observed at 16μg/ml (1×MBC) of ASP-1 treated cells after 12h of incubation (Fig. 2i) as compared to untreated cells. The majority of ASP-1 treated cells in many fields exhibited central depression or collapses with discernible holes on the cell surfaces9 (Fig. 2e, f, h and i). Irregular cluster formation of deformed S. aureus ATCC 29213 cells was observed when exposed to 1× MBC (Fig. 2f and h); this observation was consistent with the previous observation made by Greenwood and O’Grady (1972). When the effect of oxacillin (used as positive control) at 32μg/ml (1× MIC) on MRSA-4 cells was studied at different time points (Fig. 3a, d and g), the cell surface exhibited a dimpled and deformed appearance at 12h; the SEM image (Fig. 3g) also revealed a cell with a ring-like depression encircling a central perforation.
It is quite evident from Figure 3f and i that exposure to ASP-1 at 1× MBC caused damages to different extents to the MRSA-4 cell surface as evidenced in dents and pit formations. The scanning electron micrograph (Supplementary information Fig. S3a and b) at higher magnification revealed the cell surface roughness, deep perforations or holes, along with complete lysis, collapse and shrinkage of several cells. When treated with oxacillin, the SEM images (Supplementary information Fig. S4a and 4b) exhibited dimpled and deformed MRSA-4 cells at 12h with ring-like depressions encircling a central perforation (Fig. S4a).
Membrane bleb formations were also observed in the antimicrobial peptide SMAP-29-treated S. aureus cells with blebs at septum in actively dividing cells15. Some ASP-1 treated cells at 1× MBC displayed altered shapes and formation of membrane blebs as evident in Fig. 2h, and i; similar observations were made in a separate study where membrane bleb formations were observed in human defensin 5-treated Escherichiacoli5. The ultrastructure deformities at various concentrations of ASP-1 treated cells also corroborate the previous findings of the antimicrobial peptide DP-7 treated cells where membrane protuberances and plicated cell surfaces were recorded17. In a previous study, structural disorganization of phenothiazine drug methdilazine-treated S. aureus cells was reported to be similar to ASP-1 mediated ultrastructural damage4.
These observations taken together indicate that ASP-1 may be a unique antibacterial peptide exerting its effect through membrane-targeted pore formation and membrane disruption, which rarely induce drug-resistance in bacteria12. This study suggests that since the test drug had strong anti-staphylococcal and anti-MRSA activity by probably acting on the cell surface, it could present a low risk of developing drug resistance, and therefore ASP-1 might be thought as an alternative to traditional antibiotics, especially those used for the treatment of skin and mucosal infections.
Conflict of interestThe authors declare that they have no conflicts of interest.
The authors are thankful to the host University contingency. The laboratory assistance of Swetha Ramesh, a Department of Biotechnology (DBT)-sponsored Senior Research Fellow and Ms. Anviksha, a Masters in Engineering (M.E.) Biotechnology student are highly acknowledged.