Aflatoxin is a carcinogenic secondary metabolite produced mainly by Aspergillus flavus and Aspergillus parasiticus, which can seriously endanger the health of humans and animals. Oxidative stress is a common defense response, and it is known that reactive oxygen species (ROS) can induce the synthesis of a series of secondary metabolites, including aflatoxin. By using mutants lacking the afap 1 gene, the role of afap1 gene in oxidative stress and aflatoxin synthesis was assessed. The growth of the mutant strains was significantly inhibited by the increase in the concentration of H2O2, inhibition was complete at 40mmol/l. However, in the quantitative analysis by HPLC, the concentration of AFB1 increased with the increased H2O2 until 10mmol/l. Following an analysis based on the information provided by the NCBI BLAST analysis, it was assumed that Afap1, a basic leucine zipper (bZIP) transcription factor, was associated with the oxidative stress in this fungus. Treatment with 5mmol/l H2O2 completely inhibited the growth of the mutant strains in afap 1 but did not affect the growth of the CA14PTs strain (non-mutant strain). In addition, the concentration of AFB1 in the mutant strains was approximately ¼ of that observed in the CA14PTs strain. These results suggested that Afap1 plays a key role in the regulation of oxidative stress and aflatoxin production in A. flavus.
La aflatoxina es un metabolito secundario cancerígeno producido principalmente por Aspergillus flavus y Aspergillus parasiticus, que pone en riesgo grave a la salud de los humanos y los animales. El estrés oxidativo es una respuesta de defensa común, y es sabido que las especies reactivas de oxígeno (ROS) pueden inducir la síntesis de una serie de metabolitos secundarios, incluida la aflatoxina. Empleando mutantes carentes del gen afap1 se evaluó el papel de Afap1 en el estrés oxidativo y la síntesis de aflatoxinas. El crecimiento de las cepas mutadas se vio significativamente inhibido con el aumento de la concentración de H2O2, la inhibición fue completa a 40mmol/l. Sin embargo, en el análisis cuantitativo por HPLC, la concentración de la aflatoxina AFB1 aumentó con el aumento de la concentración de H2O2 hasta 10mmol/l. Tras un análisis apoyado en la información provista por la herramienta NCBI BLAST, se supuso que Afap1, un factor de transcripción de la cremallera de leucina básica (bZIP), estaba asociado con el estrés oxidativo en este hongo. El tratamiento con 5mmol/l de H2O2 inhibió completamente el crecimiento de las cepas mutantes en afap1, pero no afectó el crecimiento de la cepa CA14PTs (cepa no mutada). Además, la concentración de AFB1 en las cepas mutadas fue de aproximadamente 1/4 de la observada en CA14PTs. Estos resultados sugieren que Afap1 juega un papel clave en la regulación del estrés oxidativo y la producción de aflatoxinas en A. flavus.
Aflatoxin is a secondary metabolite produced mainly by Aspergillus flavus and Aspergillus parasiticus. As the most potent naturally-occurring toxic and carcinogenic substance, aflatoxin causes an estimated 28% of hepatocellular carcinoma (HCC), while HCC is the most common form of liver cancer in the world39 and the case rate is very high in sub-Saharan Africa, the Western Pacific region and Southeast Asia, as well as in Central America. Individuals with liver damage due to hepatitis B virus (HBV) infection are particularly vulnerable to aflatoxin invasion14. In addition, aflatoxin can lead to dysfunction of the immune system, dysplasia in children, and even death due to acute poisoning10,18. Aflatoxin contamination occurs in a wide range of food and feed commodities, including wheat, maize, peanuts, rice, peanut oil, cotton seed, milk, nuts and dairy products2. Therefore, aflatoxin not only poses a serious threat to human and animal health, but also causes huge economic losses.
The gene cluster involved in aflatoxin biosynthesis has been identified3,5,9,13,22,41,44. Most gene functions have been clarified1,9. AflR and AflS are two key transcription factors. The aflatoxin regulatory gene aflR activates the transcription of other structural genes in the aflatoxin biosynthesis pathway by encoding a positive regulatory factor6. AflS is adjacent to AflR and participates in the regulation of aflatoxin biosynthesis together with AflR. The combination of AflS and AflR forms a complex, which is then bound together in the promoter region of each structural gene in the cluster7. In addition to aflR and aflS, there are many regulatory genes involved in aflatoxin biosynthesis regulation outside the aflatoxin gene cluster. laeA and veA encode global transcription factors that regulate the biosynthesis of many secondary metabolites, such as aflatoxin, sterigmatocystin, and penicillin4,11,12,19.
There is extensive evidence that secondary metabolism is associated with oxidative stress in filamentous fungi and plants16,27,36. Based on this view, different oxidative stimuli, such as peroxides and diamide, can activate a variety of transcription factors, and many transcription factors have been proved to be involved in regulating secondary metabolism in yeast, fungi and plants. Within this network, the well-known Ap-1 transcription factor Yap-1 participated in the cellular response to oxidative stress signal in Saccharomyces cerevisiae25,33.
Like the Yap-1 roles in yeast26, several Yap-1 homologue transcription factors have been identified in filamentous fungi, and they are usually associated with resistance to H2O2 or antifungals. In the rice blast fungus Magnaporthe oryzae, Moap1 mediates the oxidative stress response and is necessary for conidia formation, apical growth and pathogenicity15. Afyap1 in Aspergillus fumigates was found to be associated with tolerance to oxidative stress29. NapA and RsmA affect stress response, sexual development and secondary metabolism in Aspergillusnidulans42. In Aspergillus ochraceus, Aoyap1 not only participated in the oxidative stress response, but also regulated ochratoxin A biosynthesis. Similarly, to Aoyap1, ApyapA in A. parasiticus also participated in the oxidative stress response and in the modulation of aflatoxin biosynthesis30. These findings suggested a probable similar link between the oxidative stress response and mycotoxin biosynthesis. However, under the oxidative stress condition, the mechanism of yap-1 homologue gene in the regulation of aflatoxin biosynthesis in A. flavus is not clear.
In this paper, afap1, the homologue of yap-1, was suggested to encode protein containing conserved bZIP domains based on the NCBI BLAST analysis. We engineered genetically modified strains of A. flavus lacking afap1 and showed the key role played by afap1 in response to oxidative stress and in the regulation of aflatoxin biosynthesis.
Materials and methodsStrains and growth conditionsThe toxigenic A. flavus CA14PTs (Δku70, ΔniaD) and recipient (Δku70, ΔniaD, ΔpyrG) strains were obtained from Dr. Perng Kuang Chang, United States Department of Agriculture, New Orleans, USA. The strain A. nidulans WJAO1 was obtained from Prof. Shihua Wang, Fujian Agriculture and Forestry University, Fuzhou, China.
Strains were activated on potato dextrose agar (PDA) plates (20g/l dextrose, 200g/l peeled potatoes and 20g/l agar) at 28°C in the dark for 3 days for conidia production. Conidial suspensions were collected from sporulated cultures of fungi on PDA plates by surface washing with sterile deionized water containing 0.1% Tween-20. The number of conidia in the suspensions was counted using a hemocytometer and diluted to 106CFU/ml with 0.1% Tween-20 solution. Conidia were cultivated in 50ml YES medium (150g/l sucrose, 20g/l yeast extract and 1g/l MgSO4·7H2O and solid medium supplemented with 16g/l agar) and grown at 28°C on a rotary incubator in the dark for AFB1 concentration detection and mycelia collection. The recipient strain was grown in broth containing yeast, glucose, trace element solution, uracil, uridine (YGTUU) (20g/l glucose, 5g/l yeast extract, 1ml trace element solution per liter of medium, 1g/l uracil and 1g/l uridine and solid medium supplemented with 15g/l agar) at 28°C for mycelial growth and conidia production. Czapek-Dox medium (Difco) supplemented with 3% sucrose was used for mutant selection.
Hydrogen peroxide sensitivity analysisFive microliters of conidia (106CFU/ml medium) from each strain were incubated in YES solid medium supplemented with different concentrations of H2O2 (0, 5, 10, 20, and 40mmol/l) for the oxidative stress response assay. All plates were cultivated in the dark at 28°C for 5 days, and colonies were photographed. Meanwhile, 5ml of conidial suspension (106CFU/ml medium) was cultured in 50ml YES liquid medium supplemented with the same concentrations of H2O2 for AFB1 concentration and the mycelial dry weight analysis. All experiments were performed in triplicate in three independent experiments.
AFB1 concentration and fungi mycelial dry weight analysisMycelia were collected and mycelial dry weight was measured after drying in a dryer (HASUC, Inc., Shanghai, China) at 65°C for 72h. AFB1 concentration in the culture filtrate was determined by extracting metabolites from the filtrate using methanol, followed by purification using an immunoaffinity column (Romer Labs, Inc., Tulln, Austria) according to the manufacturer's instructions. AFB1 concentration was detected by high-performance liquid chromatography (HPLC; Agilent Series 1260; Agilent Technologies, Santa Clara, CA, USA). HPLC was performed on an Agilent C18 Zorbax XDB column (150mm×4.6mm×5mm, Agilent Technologies), and detection was performed using a fluorescence detector (Agilent 1260; Agilent Technologies) with an excitation wavelength of 360nm and an emission wavelength of 440nm at 30°C. The mobile phase consisted of methanol/H2O (7:3, v/v) injected at a flow rate of 1ml/min.
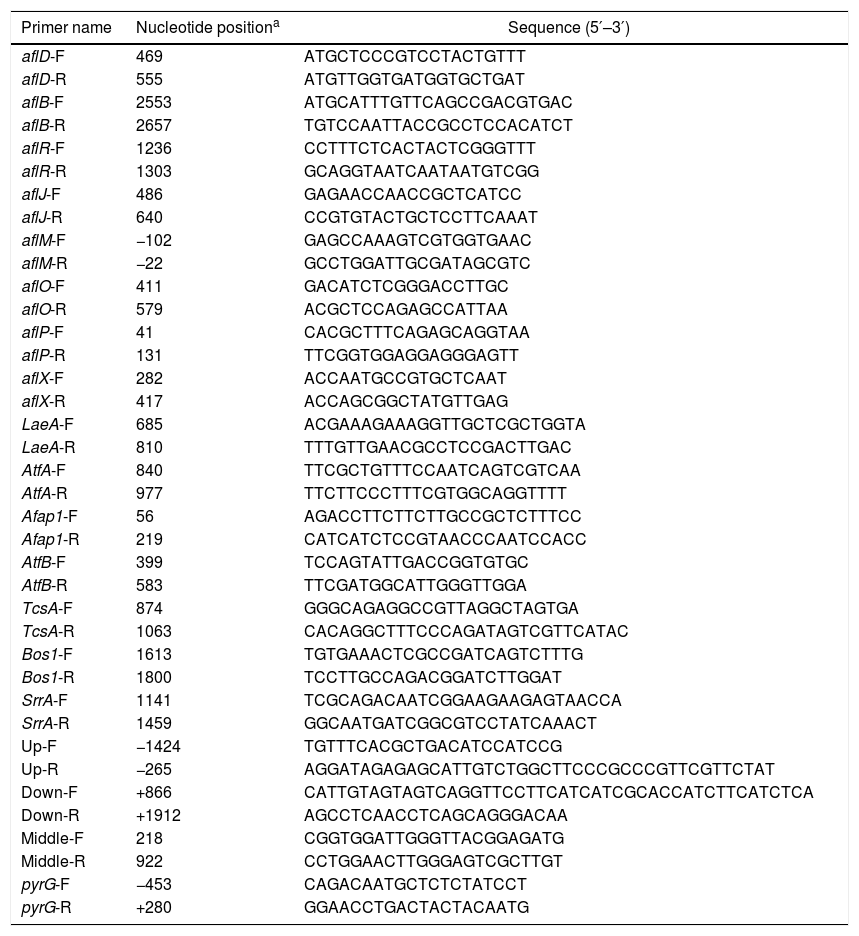
RNA extraction and quantification of gene expression by quantitative real-time PCR (qRT-PCR)Total RNA was extracted from fungal mycelia collected from YES liquid medium using a RNeasy mini kit (Qiagen, Germany) according to the manufacturer's instructions. RNA samples were treated with DNA-free DNase. The purity and concentrations of RNA were determined by measuring the absorbance of samples at 260 and 280nm using spectrophotometric quantification in a Beckman DU800 spectrophotometer (Beckman, USA). qRT-PCR was carried out in triplicate in 20-μl volumes using Power SYBR green master mix (Applied Biosystems, USA) in an ABI 7500 Real-Time PCR System (Applied Biosystems, USA). The thermal-cycling program was set as follows: 95°C for 5min, followed by 40 cycles of 95°C for 25s, 55°C to 60°C for 25s (optimized for each primer pair), and 72°C for 35s, with a melting-curve stage at 95°C for 15s, 60°C for 1min, 95°C for 30s, and 80 cycles of 60°C for 15s. Gene-specific primers were designed for each target gene using Primer 5.0 (http://www.premierbiosoft.com/primerdesign/) (Table 1). As an endogenous control, primers 18S-F (5′-GCTCTTTTGGGTCTCGTAATTGG-3′) and 18S-R (5′-CGCTATTGGAGCTGGAATTACC-3′) were used based on previous studies in order to cover 154bp of the 18S RNA gene21. Samples from each of the three biological replicates were assayed in triplicate, and data were analyzed using the ABI 7500 SDS program (Applied Biosystems, USA) by the 2−ΔΔCt method.
Primers used in this study
| Primer name | Nucleotide positiona | Sequence (5′–3′) |
|---|---|---|
| aflD-F | 469 | ATGCTCCCGTCCTACTGTTT |
| aflD-R | 555 | ATGTTGGTGATGGTGCTGAT |
| aflB-F | 2553 | ATGCATTTGTTCAGCCGACGTGAC |
| aflB-R | 2657 | TGTCCAATTACCGCCTCCACATCT |
| aflR-F | 1236 | CCTTTCTCACTACTCGGGTTT |
| aflR-R | 1303 | GCAGGTAATCAATAATGTCGG |
| aflJ-F | 486 | GAGAACCAACCGCTCATCC |
| aflJ-R | 640 | CCGTGTACTGCTCCTTCAAAT |
| aflM-F | −102 | GAGCCAAAGTCGTGGTGAAC |
| aflM-R | −22 | GCCTGGATTGCGATAGCGTC |
| aflO-F | 411 | GACATCTCGGGACCTTGC |
| aflO-R | 579 | ACGCTCCAGAGCCATTAA |
| aflP-F | 41 | CACGCTTTCAGAGCAGGTAA |
| aflP-R | 131 | TTCGGTGGAGGAGGGAGTT |
| aflX-F | 282 | ACCAATGCCGTGCTCAAT |
| aflX-R | 417 | ACCAGCGGCTATGTTGAG |
| LaeA-F | 685 | ACGAAAGAAAGGTTGCTCGCTGGTA |
| LaeA-R | 810 | TTTGTTGAACGCCTCCGACTTGAC |
| AtfA-F | 840 | TTCGCTGTTTCCAATCAGTCGTCAA |
| AtfA-R | 977 | TTCTTCCCTTTCGTGGCAGGTTTT |
| Afap1-F | 56 | AGACCTTCTTCTTGCCGCTCTTTCC |
| Afap1-R | 219 | CATCATCTCCGTAACCCAATCCACC |
| AtfB-F | 399 | TCCAGTATTGACCGGTGTGC |
| AtfB-R | 583 | TTCGATGGCATTGGGTTGGA |
| TcsA-F | 874 | GGGCAGAGGCCGTTAGGCTAGTGA |
| TcsA-R | 1063 | CACAGGCTTTCCCAGATAGTCGTTCATAC |
| Bos1-F | 1613 | TGTGAAACTCGCCGATCAGTCTTTG |
| Bos1-R | 1800 | TCCTTGCCAGACGGATCTTGGAT |
| SrrA-F | 1141 | TCGCAGACAATCGGAAGAAGAGTAACCA |
| SrrA-R | 1459 | GGCAATGATCGGCGTCCTATCAAACT |
| Up-F | −1424 | TGTTTCACGCTGACATCCATCCG |
| Up-R | −265 | AGGATAGAGAGCATTGTCTGGCTTCCCGCCCGTTCGTTCTAT |
| Down-F | +866 | CATTGTAGTAGTCAGGTTCCTTCATCATCGCACCATCTTCATCTCA |
| Down-R | +1912 | AGCCTCAACCTCAGCAGGGACAA |
| Middle-F | 218 | CGGTGGATTGGGTTACGGAGATG |
| Middle-R | 922 | CCTGGAACTTGGGAGTCGCTTGT |
| pyrG-F | −453 | CAGACAATGCTCTCTATCCT |
| pyrG-R | +280 | GGAACCTGACTACTACAATG |
The sequence of Afap1 and its homologues in S. cerevisiae, A. parasiticus, A. nidulans and Aspergillus fumigatus were used as input for BLAST in the National Center for Biotechnology Information (NCBI) database (https://blast.ncbi.nlm.nih.gov/Blast.cgi), to identify sequences with high similarities in the translated genome of A. flavus. Multiple sequence alignments were carried out using DNAssist 2.2.
Construction of the mutant strainThe deletion-mutant strain (Δafap1) was constructed as previously described8,37. A homologous transformation system for A. flavus with the pyrG gene as selection marker was used in this study35. The pyrG gene encodes an orotidine-5′-phosphate decarboxylase, which is a key gene in the synthesis of uracil nucleotides. The recipient strain CA14PTs (Δku70, ΔniaD, ΔpyrG) with the pyrG deletion cannot grow on the transformant selection medium Czapek-Dox without adding uracil and uridine, while homologous transformants carrying the pyrG gene instead of afap1 could survive on the selection medium. The detailed procedure is as follows: Genomic DNA was extracted from mycelia grown for 5 days in 50ml of YES liquid medium using benzyl chloride38,45. For the homologous fragments, the 5′ and 3′ regions of afap1 (1159 and 1046bp, respectively) were amplified with specific primer pairs Up-F/R and Down-F/R (Table 1), which contain sequences that overlap the marker gene, and were verified by sequencing. The 1600bp pyrG gene was amplified with primer pairs pyrG-F/R from A. nidulans WJAO1 genomic DNA. The PCR-fusion product was constructed and transformed into recipient strain protoplasts using polyethylene glycol buffer (15mM KCl, 20 mM CaCl2 and 1M Tris-HCl buffer, pH 7.5), and 500g/l PEG 4000. The cell suspension was plated on Czapek-Dox medium at 28°C in the dark for 5 days. Putative mutants were confirmed by PCR using primer pairs Middle-F/R and Up-F/pyrG-R and sequencing analysis.
Southern blotTransformants that passed the PCR pre-screening were further checked by Southern blot analysis, using the DIG system (Roche, Germany) in accordance with a previously described protocol24. Ten μg genomic DNA was digested with HindIII (Takara, Japan), and then electrophoresed on a 1% agarose gel to separate by size. A sheet of nylon membranes (Hybond N+, Pharmacia, USA) was placed on top of the gel for DNA transference. The hybridization probe with DIG-labeled was synthesized with PCR DIG probe synthesis kit (Roche, Germany) by following the manufacturer's protocol. The probe matched the downstream sequence of the homology arm in the homologous recombination fragment was generated by PCR amplification using primers 5′-AACGTGGTTGTATTTGCCCC-3′ and 5′-GCTCTTGGACAATGCTCTCG-3′.
Statistical analysisStatistical analyses of the obtained data were performed using SPSS 21.0 software (IBM, Chicago, IL, USA). Differences between the means were evaluated by a one-way analysis of variance (ANOVA), and in all cases, statistical significance was established at p<0.05.
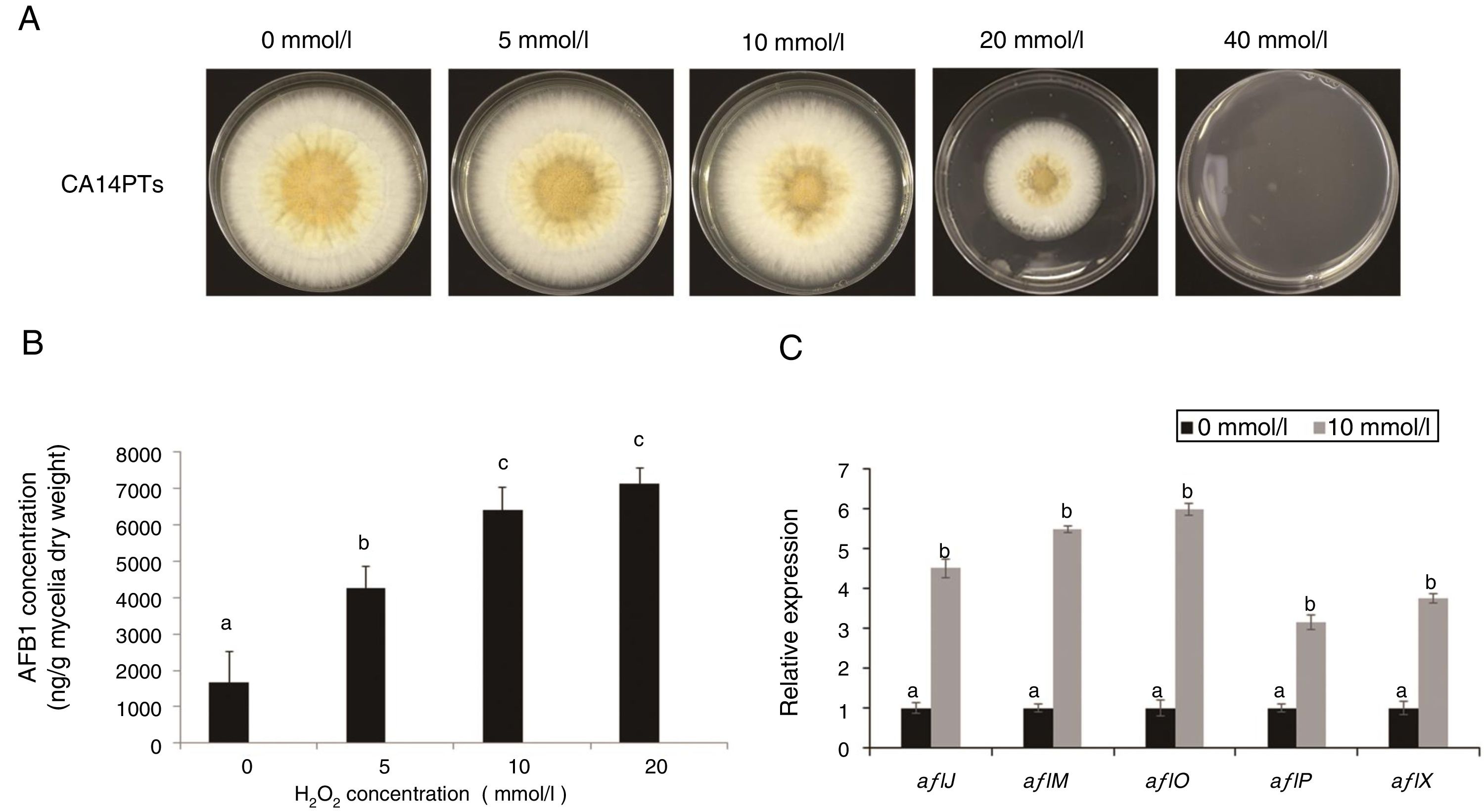
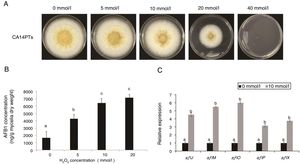
ResultsEffect of different oxidative stress on A. flavus growth and AFB1 productionOxidative stress is a very important environmental stimulus for fungi. To evaluate the effect of oxidative stress on the growth and AFB1 production of A. flavus, H2O2 solutions at different concentrations (0, 5, 10, 20, and 40mmol/l) were added to YES plates and liquid culture medium, respectively. The growth of A. flavus CA14PTs was significantly inhibited by the increased H2O2 and were completely inhibited at 40mmol/l (Fig. 1A). The AFB1 concentration in YES broth was increased after the treatment of H2O2 at the concentration of 5, 10 and 20mmol/l, respectively (Fig. 1B). There is no obvious increase for AFB1 concentration at 20mmol/l H2O2 compared with 10mmol/l. In addition, the expression levels of key aflatoxin biosynthetic structural genes (aflJ, aflM, aflO, aflP and aflX) were up-regulated by the treatment of 10mmol/l H2O2 according to qRT-PCR (Fig. 1C). The data based on the results indicated that oxidative stress could affect strain growth and stimulate aflatoxin biosynthesis.
Colony growth and aflatoxin production after 0, 5, 10, 20 and 40mmol/l H2O2 treatment. The A. flavus toxigenic strain CA14PTs was grown for 5 d at 28°C in darkness. (A) View from top of colony. (B) Determination of AFB1 concentration per unit of mycelial weight by HPLC in response to H2O2. Different letters indicate that there were statistically significant differences (p=0.05). (C) Relative expressions of aflatoxin synthesis genes. Samples were harvested after H2O2 treatment (10mmol/l), and gene expression was measured by quantitative real-time PCR. Standard errors of the mean are shown (n=3). Different letters indicate that there were statistically significant differences (p=0.05).
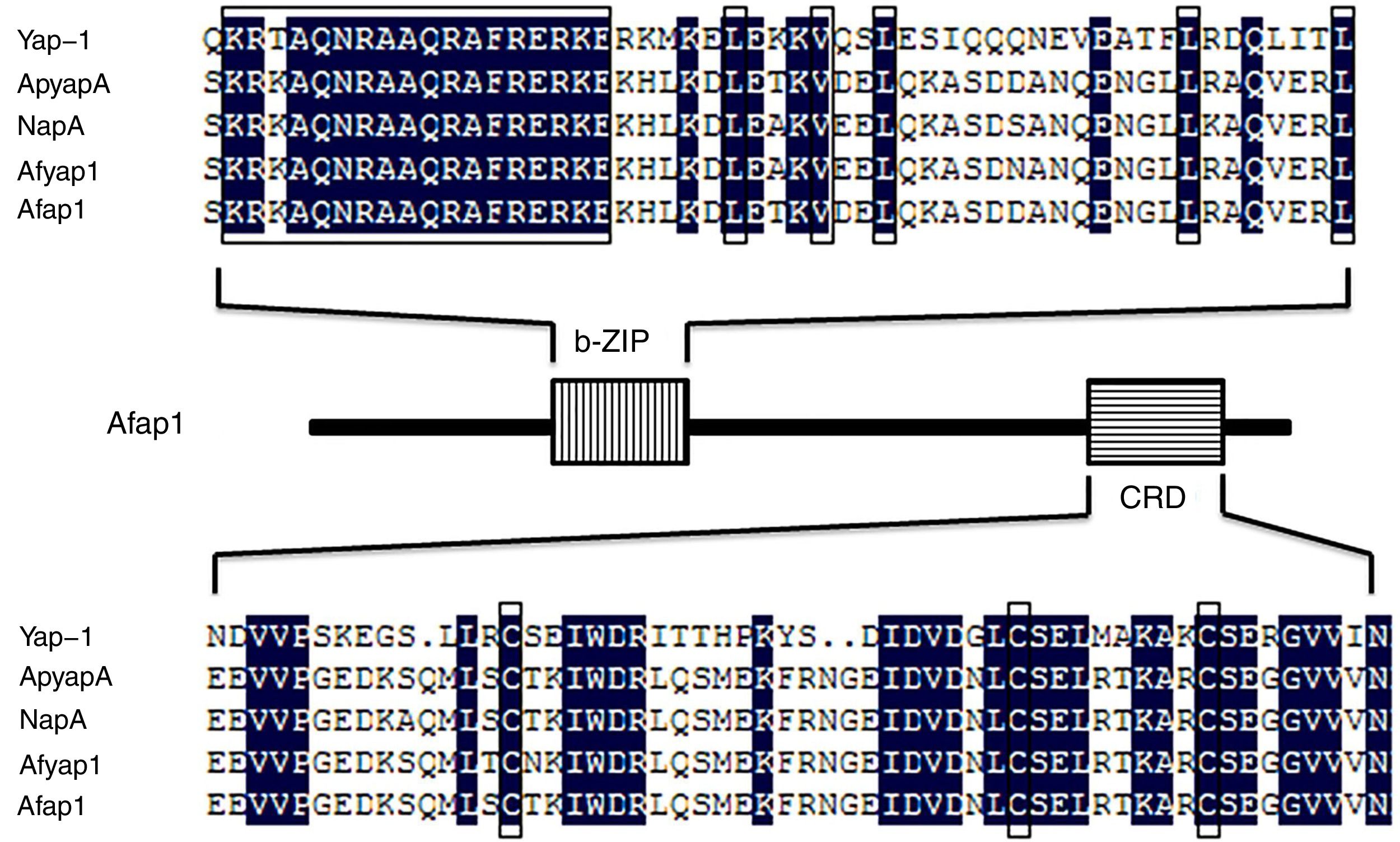
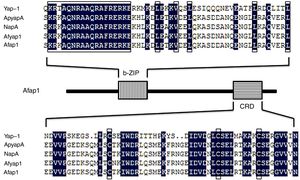
By the NCBI BLAST analysis, Afap1 was identified as a putative bZIP transcription factor. Alignment of the Afap1 protein sequence to those of Yap-1 (S. cerevisiae S288c), ApyapA (A. parasiticus SU-1), NapA (A. nidulans FGSC A4) and Afyap1 (A. fumigatus Af293) (Fig. 2) showed two conserved domains: A C-terminal nuclear export signal (NES) embedded in a characteristic cysteine-rich domain (c-CRD) and a N-terminal basic leucine zipper domain (bZIP domain). Afap1 has lower homology (16.22% similarity) with its yeast orthologues, but it has higher homology with other filamentous ascomycetes. The conserved bZIP domain and cystein-rich domain (CRD) suggested that Afap1 has a similar role in response to oxidative stress and toxin biosynthesis in A. flavus as other homologue proteins.
Amino acid sequence alignment of the characteristic domain. Yap-1 is from S. cerevisiae S288c, ApyapA is from A. parasiticus SU-1, NapA is from A. nidulans FGSC A4, Afyap1 is from A. fumigatus Af293, Afap1 is from A. flavus. The bZIP domain is shown as a vertical line box, and the cysteine-rich domain (CRD) as a horizontal line box. The amino acid sequence of these domains of Afap1 is aligned with those of Yap-1, ApyapA, NapA, and Afyap1. Boxes indicate conserved regions of different functional domains.
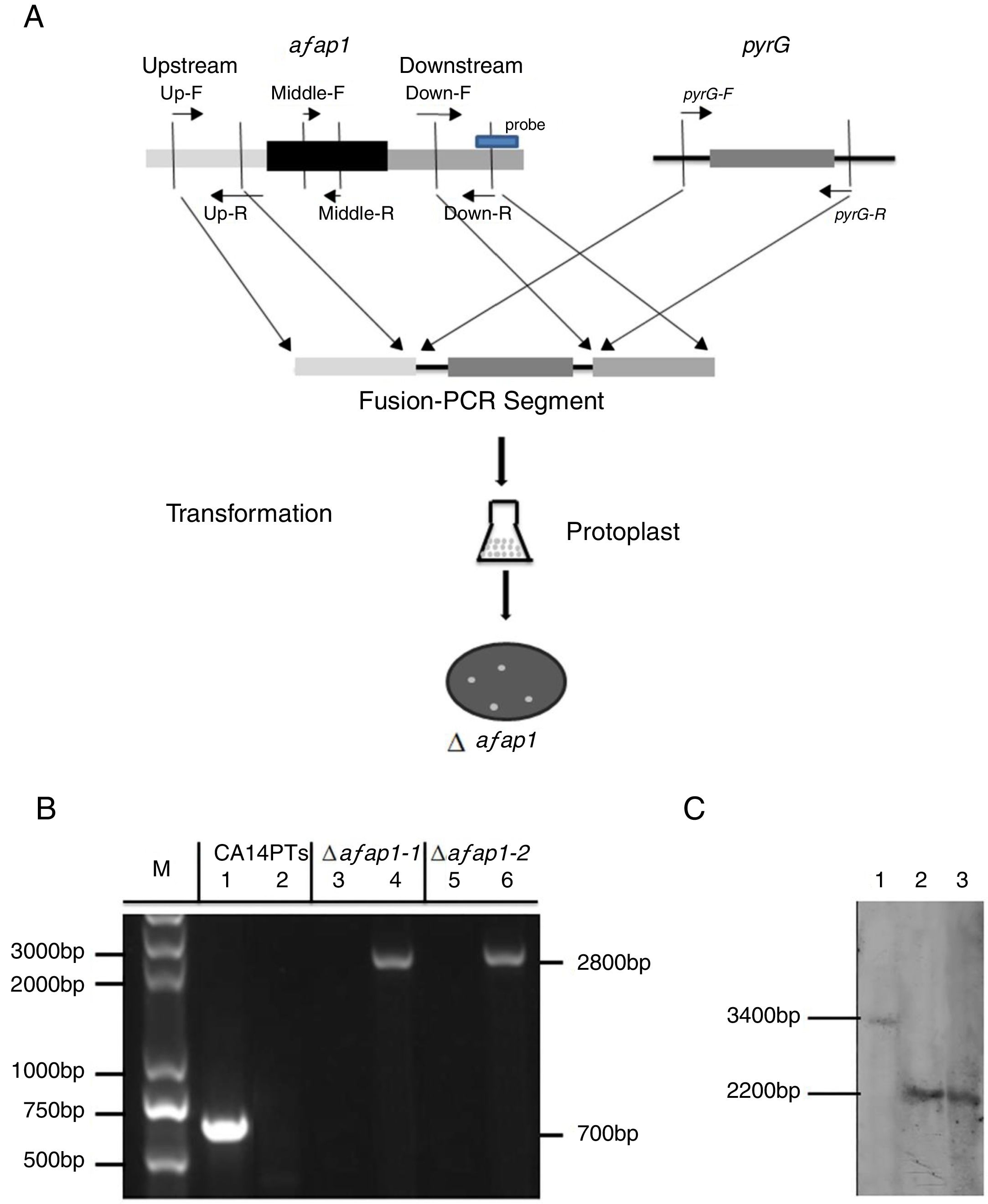
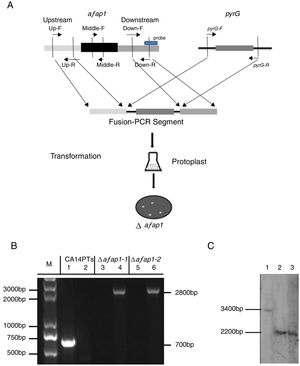
To analyze the role of afap1 in oxidative-stress response and aflatoxin biosynthesis, Δafap1 mutants were generated using homologous recombination (Fig. 3A). Two transformants (i.e., Δafap1-1 and Δafap1-2) were selected for further PCR verification. A 700bp fragment, encoding for partial ORF of afap1, could be amplified with primers Middle-F/R in CA14PTs but not in the positive transformants. A 2800bp fragment, encoding for pyrG and upstream of afap1, could only be amplified with primers Up-F/pyrG-R in positive transformants. As shown in Figure 3B, only the 2800bp fragment was observed in Δafap1-1 and Δafap1-2. Further sequencing analysis showed that the gene afap1 was exactly replaced by pyrG in these two mutants. Additionally, southern blot hybridization revealed a 3.4kb fragment and a 2.2kb fragment in CA14PTs and the afap1 mutants when digested with HindIII, respectively (Fig. 3C). It is confirmed that there are sequence differences between the afap1 mutants and CA14PTs as expected. Combined with the above homologous recombination strategy and PCR analysis, it was shown that afap1 was properly deleted and mono-copy.
Construction and verification of the afap1-deletion mutants. (A) Afap1 gene-replacement strategy. Primers are shown in Table 1. (B) PCR confirmation of the Δafap1 mutants. Lanes 1, 3 and 5 results are from the use of the Middle-F and Middle-R primers, and lanes 2, 4 and 6 results are from the use of the Up-F and pyrG-R primers. M, DNA marker; lanes 1 and 2, CA14PTs; lanes 3 and 4, Δafap1-1; lanes 5 and 6, Δafap1-2. (C) Southern blot hybridization. Lanes 1, CA14PTs; lanes 2, Δafap1-1; lanes 3, Δafap1-2. In CA14PTs, probe reveals a fragment of 3.4kb when digested with HindIII. In the mutant strains Δafap1, probe reveals a fragment of 2.2kb when digested with HindIII.
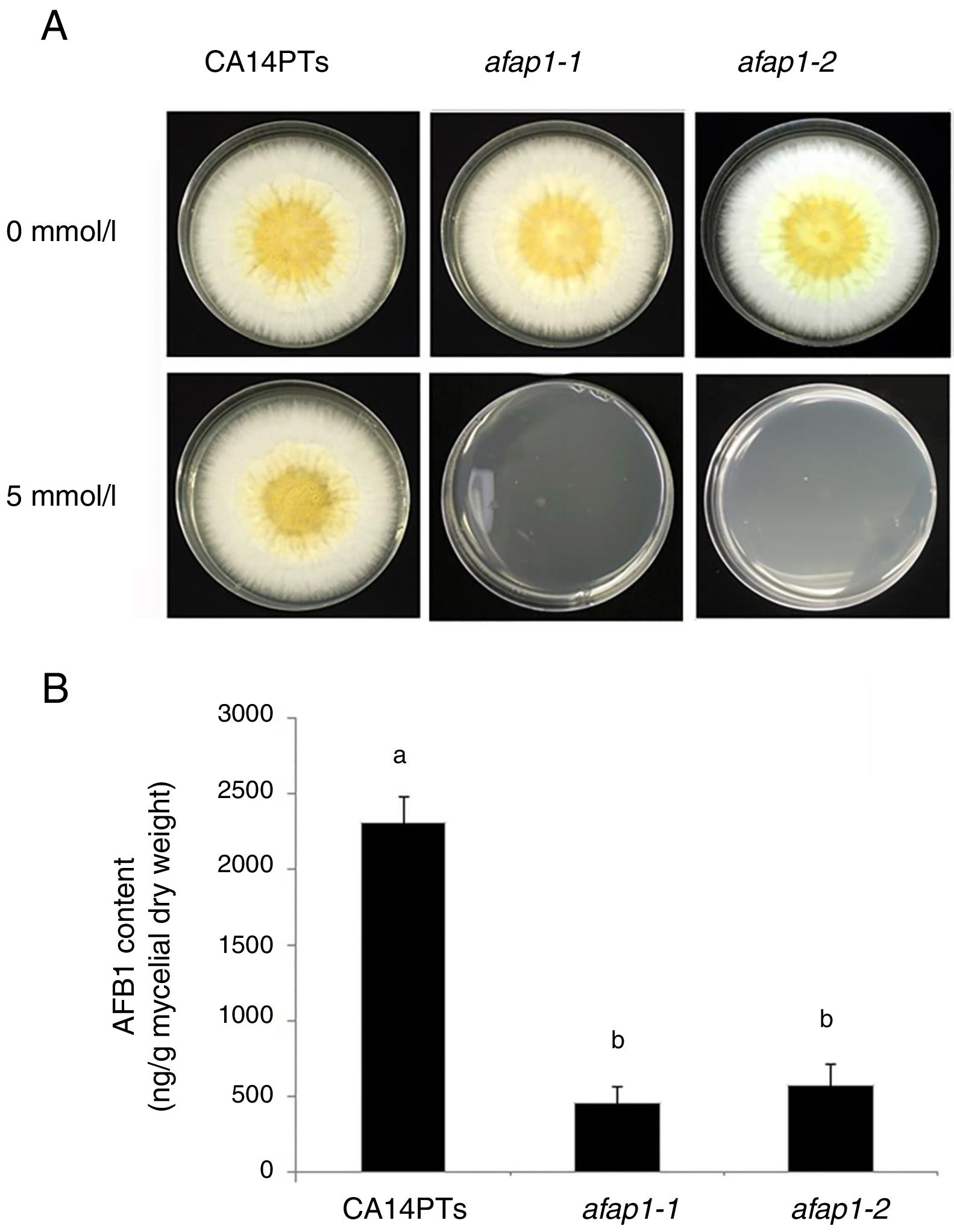
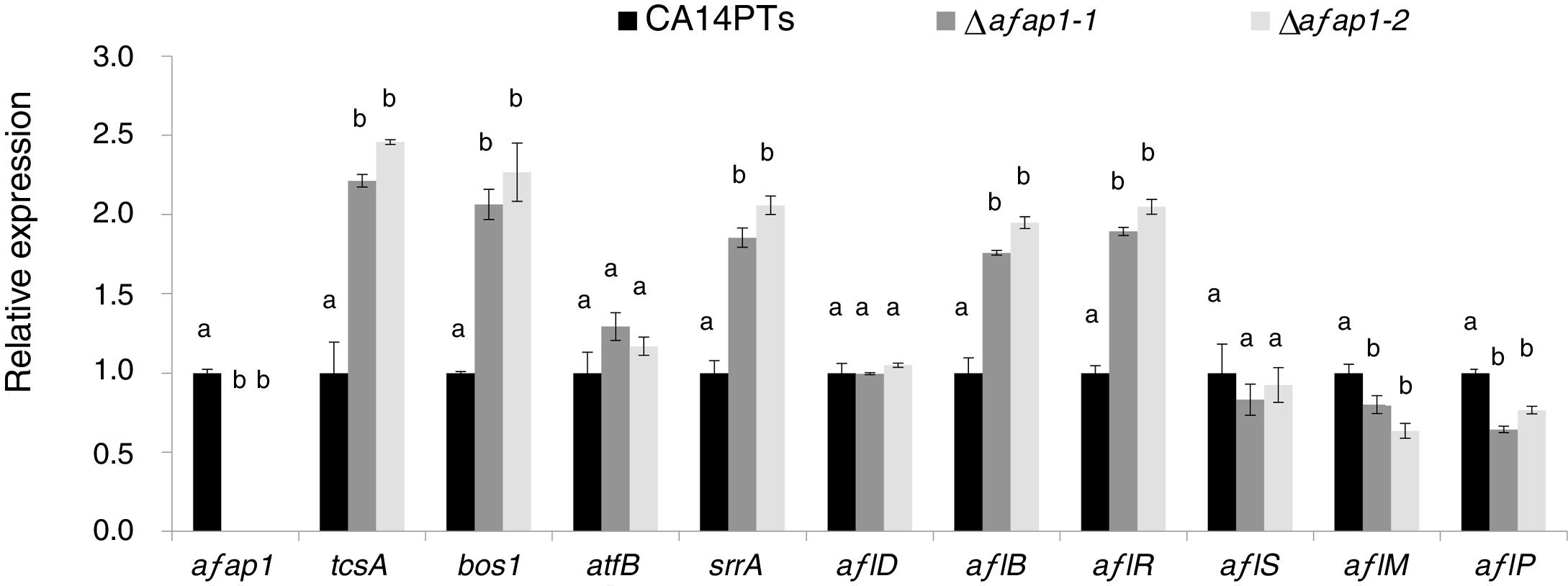
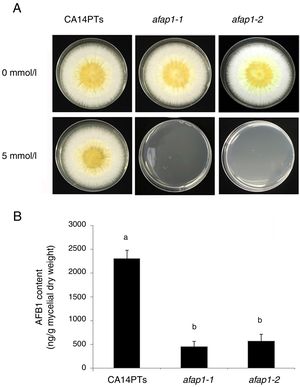
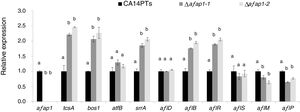
CA14PTs and the Δafap1 mutants were incubated in YES plates supplemented with different concentration of H2O2 (Fig. 4A). The growth rates of the CA14PTs and Δafap1 mutants were similar on YES plates without H2O2. However, the growth of Δafap1 mutants was completely inhibited by 5mmol/l H2O2. In contrast, the growth of CA14PTs was not inhibited even at 10mmol/l H2O2. Meanwhile, AFB1 concentration of the Δafap1 mutants was significantly decreased by around 75%, which compared to CA14PTs (p<0.01) (Fig. 4B). In addition, the expression levels of the key transcription factors genes (tcsA, bos1, atfB and srrA) related to the oxidative-stress response and aflatoxin biosynthetic genes (aflD, aflB, aflR, aflS, aflM and aflP) in YES liquid medium were detected using qRT-PCR. The expression of tcsA, bos1, srrA, aflB and aflR was significantly up-regulated in the Δafap1 mutants compared to CA14PTs, and the expression of aflM and aflP were significantly down-regulated. The expression of atfB was up-regulated in the Δafap1 mutants, although the difference was not significant (Fig. 5).
Defects of CA14PTs and Δafap1 mutants in response to different oxidative stress. (A) CA14PTs and Δafap1 mutants were separately incubated in YES medium under different oxidative-stress conditions for 5 days. (B) HPLC analyses of AFB1 concentration per unit of mycelial weight. Different letters indicate that there were statistically significant differences (p=0.05).
Quantitative real-time PCR analyses of genes related to oxidative stress and aflatoxin biosynthesis in the Δafap1 mutants as compared with those in CA14PTs. All data represent the means of three independent samples, and standard errors of the means are shown (n=3). Different letters indicate that there were statistically significant differences (p=0.05).
Oxidative stress is one of the earliest responses and a common cell defense mechanism in living things. Cellular response to oxidative stress plays a crucial role in plants, vertebrates and fungi; it enables the cell to survive a variety of extra- and intracellular oxidative stressors. The classical review of the oxidative stress response in fungi was developed based on research in yeast which showed that regulation of defense-related antioxidant genes contributed to the survival of the organism. The regulation of secondary metabolism is closely linked to the cellular response to oxidative stress in filamentous fungi and contributes to the complexity of the response16. However, this response is most complicated and robust than that of yeast in response to various environmental conditions.
Previous reports strongly suggested that several transcription factors associated with the Stress Activated Protein Kinase/Mitogen-Activated Protein Kinase (SAPK/MAPK) pathway coordinate the transcriptional level of secondary metabolism genes and antioxidant enzymes, thereby controlling the metabolic processes in cellular stress response. Ap-1 family is one of the most important transcription factors. Ap-1 family have many homologous proteins in S.cerevisiae33 and Aspergillus spp.30,31; however, its role in toxin biosynthesis and virulence is divergent28. In A. parasiticus and A. ochraceus, deletion of apyapA and aoyap1 resulted in increases of aflatoxin and ochratoxin, respectively30,32. In Fusarium graminearum, the Δfgap1 mutant showed higher sensitivity to oxidative stress (H2O2) and higher level of trichothecene concentration associated with overexpression of TRI genes. However, the activation mechanism of toxin accumulation in response to oxidative stress was not observed25. In contrast, in A. nidulans, deletion or overexpression of napA led to a decreased tolerance to oxidative stress and sterigmatocystin synthesis42. In this study, the growth of CA14PTs was significantly inhibited following treatment with 20mmol/l H2O2, whereas growth of the knockout mutants was completely inhibited following treatment with only 5mmol/l H2O2 due to the lack of a key transcription factor Afap1 related to the oxidative-stress response. The effect of deletion of afap1 is similar with napA and is contrary to aoyap1, apyapA and fgap1.
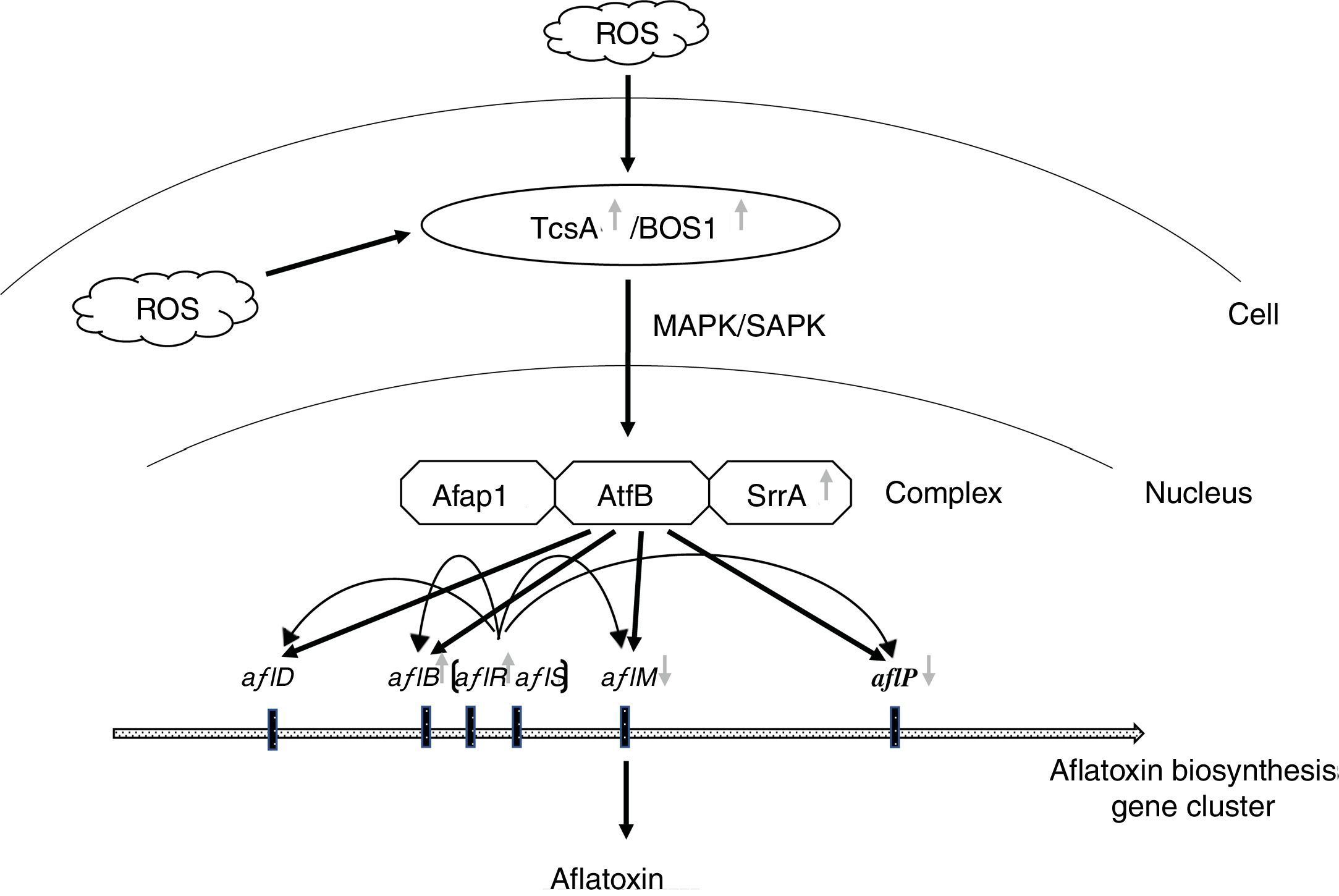
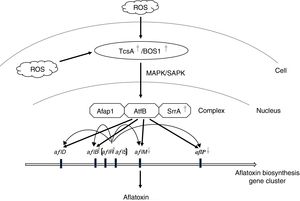
Hong et al. (2013) proposed that SrrA (SrrA recruits AP-1) and AtfB combined with the promoter regions of aflatoxin biosynthetic genes to help their induction by transcription factor AflR17. Moreover, AtfB has been proved to combine with the promoter regions of aflatoxin biosynthetic genes including aflB (fas-1), aflD (nor-1), aflM (ver-1) and aflP (omtA), which carry CRE sites17,34. In addition, the sensor kinases TcsA transmit oxidative stress signals through SrrA and/or SskA response regulators23, and then cooperate with Ap-1 against oxidative stress in other Aspergillus spp.16. Therefore, a similar pathway may have A. flavus since oxidative stress signals were transmitted through sensor kinases (ortholog of TcsA or Bos1 in yeast) to AtfB-SrrA-Afap1 homologous complex, and then induced aflatoxin biosynthesis (Fig. 6). In this study, the expression of tcsA, bos1, srrA and aflR was up-regulated in the Δafap1 mutants compared to CA14PTs. aflB, encoding fatty acid synthase and being close to aflR in the aflatoxin gene cluster, was also up-regulated in the Δafap1 mutants. However, the expression of aflM and aflP, two downstream structural genes, were significantly down-regulated in the Δafap1 mutants, which resulted in a down-regulation of aflatoxin production. On the other hand, interestingly, the gene expression of aflS was down-regulated in the knockout mutants. Down-regulation of aflS led to the decreased production of AflS protein, which is beneficial for some potential suppressors to bind to AflR in place of AflS. Consequently, the transcription of the aflatoxin biosynthesis gene, which relies on AflS-AflR, would be reduced and aflatoxin biosynthesis would decrease. There were similar findings in some previous studies20,40,43.
The present study revealed that oxidative stress inhibited the growth of toxigenic strains and was completely inhibited at 40mmol/l H2O2. However, the AFB1 concentration was increased until 10mmol/l. According to the NCBI BLAST analysis, transcription factor Afap1 has the conserved protein domains of other AP-1 homologue proteins. Deletion of afap1 resulted in an increase in sensitivity to oxidative stress and a decrease in aflatoxin production in A. flavus. These results suggested that afap1 plays a key role in tolerance to oxidative stress and promoted aflatoxin production in A. flavus.
Conflict of interestThe authors declare that they have no conflicts of interest.
We gratefully acknowledge the financial support of National Key R&D Program of China (2017YFC1600903), National Program of China Basic Science and Technology Research (2013FY113400).