The objective of this study was to identify twelve Brucella abortus isolates of bovine origin from the department of Nariño in Colombia up to the biovar level. These isolates are included in the collection of the Germplasm Bank of Microorganisms of Animal Health Interest – Bacteria and Virus (BGSA–BV). The identification was carried out through conventional methods such as macro and microscopic morphological descriptions, enzymatic activity, biochemical profile, substrate use and sensitivity to dyes. Complementary genotypic characterization was carried out using multiplex PCR for B. abortus, Brucella melitensis, Brucella ovis, and Brucella suis-Erytritol (AMOS-ERY-PCR), RFLP-IS711, by southern blot hybridization, as well as by the multiple locus variable number of tandem repeat analysis (MLVA) using the ery gene and the insertion sequence IS711 and variable number of tandem repeats (VNTR) as molecular markers. The results of the phenotypic and molecular characterization allowed to identify twelve isolates as B. abortus biovar 4 as well as to differentiate field from vaccine strains. This is the first study on the phenotypic and molecular identification of B. abortus isolates in Colombia. It was concluded that the phenotypic and molecular identification of twelve isolates as B. abortus biovar 4 could be achieved using conventional and molecular techniques with enough resolution power. The identification of these isolates to the biovar level in taxonomic and epidemiological terms will allow the use of this genetic resource as reference strains in future research. This finding constitutes the basis for identifying biotypes not previously reported in the country that might be useful to support brucellosis survey programs in Colombia.
El objetivo de este estudio fue identificar 12 aislamientos de Brucella abortus de origen bovino procedentes del departamento de Nariño, Colombia, hasta la descripción de biovar. Estos aislamientos conforman la colección del Banco de Germoplasma de Microorganismos de Interés en Salud Animal, Bacterias y Virus. La identificación se hizo mediante métodos convencionales, como la descripción morfológica macro y microscópica de actividad enzimática, de perfiles bioquímicos, de utilización de sustratos y de sensibilidad a colorantes. Se hizo una caracterización genotípica complementaria mediante PCR múltiple para Brucella abortus, Brucella melitensis, Brucella ovis y Brucella suis-eritritol (AMOS-ERY-PCR); RFLP-IS711; hibridación Southern blot y análisis multi-locus de repeticiones en tándem de número variable (MLVA), empleando como marcadores moleculares el gen ery, la secuencia de inserción IS711 y el número variable de repeticiones en tándem (VNTR). Los resultados de la caracterización fenotípica y molecular permitieron identificar 12 aislamientos de campo como B. abortus biovar 4 y diferenciar cepas de campo de cepas vacunales. Este es el primer estudio de identificación fenotípica y molecular de aislamientos de B. abortus en Colombia. Por su importancia taxonómica y epidemiológica, la identificación de estos aislamientos hasta el nivel de biovar permitirá disponer de recursos genéticos que se pueden emplear como cepas de referencia en futuras investigaciones. Estos resultados pueden considerarse como una base para la identificación de biotipos no reportados en el país y podrán ser utilizados en programas de monitoreo y vigilancia de la brucelosis bovina en Colombia.
Bovine brucellosis caused by Brucella abortus is an endemic zoonotic disease of high impact in public health and in the livestock industry that causes huge economic losses and constrains the international trade of products of animal origin.
The genus Brucella includes facultative, intracellular, Gram-negative and slightly acid-alcohol-resistant bacteria. Currently, two different groups have been identified in the genus Brucella: the classical Brucella group including B. abortus, B. melitensis, B. suis, B. ovis, B. canis, B. neotomae, B. ceti, B. pinnipediae and B. papionis and the atypical or non-classical species of Brucella including B. microti and B. inopinata36,37,40,42. In 2016 a new species named B. vulpis was isolated from mandibular lymph nodes of a wild red fox (Vulpes vulpes) in Austria36.
This classification was initially based on phenotypic characteristics such as CO2 requirements, hydrogen sulfide (H2S) production, urease activity, sensitivity to different dye concentrations, susceptibility to specific phages and host preference11,16,19,24,43. Currently, Brucella classification is completed with molecular tests, sequencing, and whole genome sequencing annotation36.
Three classical species have been subdivided into biovars as follows: eight B. abortus biovars, three B. melitensis biovars and five B. suis biovars35. Due to the high genetic homology of the species of the genus Brucella by DNA hybridization assays (higher than 90%), the fact that there is sequence similarity higher than 98% by comparative genomics and the complete sequence identity in the 16 rRNA gene among all the Brucella species, Bardenstein et al.4 and Scholl et al.26 proposed that the species of this genus should be described as biovars of the species B. melitensis. However, the molecular studies have demonstrated that the classical species correspond to genetically distinct species besides being narrowly related5.
Two molecular markers have been used to identify the genus, the species and the differentiation of Brucella biovars. On the one hand, some genes involved in erythritol catabolism (a compound used by all members of the Brucella genus as a source of carbon) and on the other hand, the insertion sequences in the genome of this microorganism8,29,30,39. A promissory molecular strategy to differentiate between species and biovars of Brucella is the polymorphism based on the distribution and number of copies of the IS711 insertion sequence in the Brucella genome22,23. The stability and polymorphism of IS711 are used to differentiate B. abortus biovars 1, 2, and 4 from biovars 3, 5, and 9. To differentiate B. abortus, B. melitensis biovars 1, 2 and 3, B. ovis and B. suis biotype 1, a multiple PCR called AMOS-PCR is used to amplify fragments whose sizes are distinctive for each species8. This test was later improved in order to differentiate between field and vaccine strains in brucellosis eradication programs and was called AMOS-ERY-PCR28. However, this technique has the disadvantage of not having enough resolution to identify biovars of the same species of B. abortus, e.g. biovars 1, 2 and 430.
Currently, the publication of the complete genomes of several Brucella species has allowed to search for hypervariable DNA sequence regions that are useful as molecular markers to be able to diagnose, identify and differentiate between species and biovars. Among these markers there are microsatellites or regions that correspond to the variable number of tandem repeats (VNTR), which have shown to be highly polymorphic genetic markers that are useful for the differentiation of pathogens that have little genomic diversity such as Brucella spp.3,18,35.
Moreover, the analysis of various loci or VNTR regions is called multiple locus variable number of tandem repeats analysis (MLVA). Several MLVA schemes for Brucella spp. have been published. These schemes use various VNTR loci that include markers with different levels of diversity and discriminant power5,14,17,38,41. The MLVA molecular typification system evaluates various VNTR regions that are achieved through the individual or multiple PCR amplification of these loci by using specific primers for the VNTR flanking regions. The genetic profile and the size of the amplified fragments are established by electrophoresis in agarose gels, easily comparable to patterns obtained from reference strains; this facilitates its use and interpretation; therefore, the MLVA becomes a quick, simple and economic Brucella biovar typification method32. The multilocus nature of these VNTR sequences gives a precise vision of the frequency and geographic distribution of biovars and genetic variants of B. abortus. This is an essential requirement to trace the dissemination routes of the pathogen27.
Therefore, the objective of this study was to classify the Colombian B. abortus isolates that are kept in the Germplasm Bank of Microorganisms of Animal Health Interest – Bacteria and Viruses (BGSA–BV, for its acronym in Spanish) to the biovar level using phenotypic and molecular tests. The characterization of these isolates might be useful in other biovar epidemiological distribution studies of B. abortus in Colombia.
Materials and methodsB. abortus strains and reference materialThe isolates of this study were obtained from the bacteria and viruses collection belonging to the Germ Bank of Microorganisms of Animal Health Interest – Bacteria and viruses (BGSA–BV) from the Colombian Corporation for Research in Agriculture (AGROSAVIA) (Table 1). Two vaccine strains were used as reference material: B. abortus RB51* biovar 1 (Schering-Plough) and B. abortus strain 19 biovar 1 (VECOL), as well as four reference strains: B. abortus biovar 2, B. abortus biovar 3, B. abortus biovar 4, and B. abortus biovar 9, which had been donated in 1998 by the former Pan American Zoonoses Center (CEPANZO) and are kept at −80°C as part of the BGSA-BV. In the AMOS-PCR and MLVA-2 strategy, genomic DNA from Salmonella enterica serovar Enteritidis (ATCC 13076) Staphylococcus aureus (ATCC 25923) was used.
Description of the Brucella abortus isolates used in this study
| Isolates | Sample | Location (municipalities)* of isolation | Year of isolation |
|---|---|---|---|
| Ba-Col-B-001 | Placenta | Cumbal | 1997 |
| Ba-Col-B-002 | Vaginal fluids | Cumbal | 1997 |
| Ba-Col-B-003 | Fetal abomasum content | Pasto | 1996 |
| Ba-Col-B-004 | Fetal abomasum content | Pasto | 1996 |
| Ba-Col-B-005 | Fetal abomasum content | Pasto | 1996 |
| Ba-Col-B-006 | Fetal abomasum content | Pasto | 1997 |
| Ba-Col-B-007 | Fetal abomasum content | Guachucal | 1997 |
| Ba-Col-B-008 | Fetal abomasum content | Guachucal | 1996 |
| Ba-Col-B-009 | Fetal abomasum content | Túquerres | 1998 |
| Ba-Col-B-010 | Placenta | Ipiales | 1997 |
| Ba-Col-B-011 | Vaginal fluids | Ipiales | 1997 |
| Ba-Col-B-012 | Vaginal fluids | Pupiales | 1997 |
The identification of isolates to species and to biovar levels was carried out by conventional methods such as CO2 requirements, hydrogen sulfide production on strips of lead acetate, semiquantitative urease enzyme production for 1–2h of incubation, the cytochrome-oxidase test, the slide agglutination test with anti-Brucella abortus polyclonal serum (Difco), growth in the presence of erythritol sugar at a range of 1–2mg/ml, growth in the presence of dyes such as thionine (1:5000 and 1:10000 dilutions), basic fuchsine (1:25000 and 1:50000 dilutions) and safranin (1:5000 and 1:10000 dilutions)17,24,28,44. Cellular morphology was established through the Gram stain technique and the Ziehl–Neelsen or Stamp's modified staining methods12,43.
Molecular identification of B. abortus isolatesDNA extractionIsolates of B. abortus were cultivated in BHI agar for molecular identification. After an incubation period of 48h in a 5% CO2 atmosphere, bacteria were suspended and inactivated by heat. Genomic DNA was extracted through the CTAB-phenol chloroform method, precipitation with ethanol for the AMOS-ERY-PCR, and IS711-RFLP-Southern blot method. For the RFLP-Southern blot a DNA concentration of 400ng/μl was used8,30. For the MLVA technique the Brucella spp. cultures were boiled for 10min in T.E. buffer, centrifuged at 12000×g, 5min and then the supernatant was used as a DNA template25,32. All DNA samples were preserved at −20°C until they were used.
AMOS-ERY-PCRA cocktail of seven original oligonucleotides was prepared as follows: IS711 that hybridizes with the IS711 insertion sequence; four oligonucleotides PBa, PBm, PBo, PBs that hybridize inside the alkB gene in variable distances of IS711 generating amplicons of different species-specific sizes8,17,23, and two oligonucleotides, ERY1 and ERY2 that hybridize with a region of the ery gene which is common to all the Brucella spp. strains, except to vaccine strain 1930 (Table 2). PCR was carried out according to the protocol established by Ocampo et al.30 as follows: a volume of 25μl composed by 0.3mM of dNTPs (Invitrogen®), 1× PCR buffer (Invitrogen®), 1.5mM of MgCl2 (Invitrogen®), 0.2μM of each primer 1U of Taq polymerase (Invitrogen®) and 30ng/μl of genomic DNA. The amplification conditions were: initial denaturation at 94°C for 10min followed by 30 cycles of denaturation at 94°C for 30s, annealing at 60°C for 30s, extension at 72°C for 2min and a final extension at 72°C for 5min.
Probes and primer sequences used in AMOS-ERY-PCR and HOOF-Prints-MLVA-2
| Technique | Primers | Sequences 5′–3′ | Size bp | Reference |
|---|---|---|---|---|
| AMOS-ERY-PCR | PBa | GACGAACGGAATTTTTCCAATCCC | 492 | [8] |
| PBm | AAATCGCGTCCTTGCTGGTCTGA | 731 | [8] | |
| PBo | CGGGTTCTGGCACCATCGTCG | 796 | [8] | |
| PBs | GCGCGGTTTTCTGAAGGTTCAGG | 285 | [8] | |
| IS711 | TGCCGATCACTTAAGGGCCTTCAT | [8] | ||
| ERY1 | CGCCTGCGTGACCTCCAGCTTACCC | 1270 | [31] | |
| ERY2 | GGCCATGACACGCGGCATATAACC | |||
| HOOF-Prints-MLVA-2 | LOCUS-1 | GGTGATTGCCGCGTGGTTCCGTTGAATGAG | 144–193 | [7] |
| REV-3 | GGGGGCARTARGGCAGTATGTTAAGGGAATAGGG | |||
| LOCUS-7 | CAGAGCCGTCGGTGGTTACTTGAGTAGGGCAG | 91–157 | [7] | |
| REV-1 | GGGGAGTATGTTTTGGTTGCGCATGACCGC |
The restriction profiles of the IS711 insertion sequence were carried out according to the protocol described previously by Bricker and Halling8 and Ocampo et al.30 with some modifications: the probe of approximately 700bp containing the IS711 genetic element was generated by PCR and marked with digoxigenin (DIG) using the PCR DIG Probe Synthesis Kit (Roche Diagnostics GmbH) following the manufacturer's instructions. The genomic DNA of the B. abortus 19 vaccine strain was employed as a mold. PBa and IS711 primers were used in the probe construction; the PCR amplification conditions were the same as the ones employed in AMOS-ERY-PCR.
For the Southern blot hybridization technique, the DNA of the isolates was digested with the EcoRI (Invitrogen®) restriction enzyme, then the restriction fragments were resolved in agarose gel at 1% and were transferred to nylon membranes by capillarity (Roche Diagnostics GmbH). The DNA transferred was hybridized with a DIG-labeled probe, the hybridization signal was revealed with chemiluminescence using anti-DIG-alkaline phosphatase and CDP-Star chemiluminescent substrate (Roche Diagnostics GmbH) following the manufacturer's instructions. The hybridization signal was registered in chemiluminescent detection film during a 15-min exposure period at room temperature (Roche Diagnostics GmbH). To establish the IS711-RFLP profiles the film images were captured by the Gel Doc EZ™ Bio-Rad photodocumentation system.
Multiple locus variable number of tandem repeat analysis (MLVA)The B. abortus isolates were analyzed by the MLVA technique based on the amplification of two of the eight loci (locus 1 and locus 7) described in HOOF-Prints or hypervariable octameric oligonucleotide finger-print technique6,7 (Table 2). These two VNTR loci were chosen because they show a high allelic diversity between biovars and a high discriminant power between them, according to Bricker and Ewalt6, Higgins et al.14 and Hollender et al.15
A multiplex PCR protocol named MLVA-2 and based on the procedure described by Bricker et al.7 was standardized. The procedure is as follows: a volume of 50μl composed by 0.25mM of dNTPs (Invitrogen®), 1× PCR buffer (Invitrogen®), 2.5mM of MgCl2 (Invitrogen®), 0.2μM of each primer, 0.6U of Taq polymerase (Invitrogen®) and 50ng/μl of genomic DNA. The amplification conditions were: initial denaturation at 94°C for 2min and 32 cycles of denaturation at 94°C for 15s, annealing at 60°C for 20s, extension at 72°C for 1.5min and a final extension at 72°C for 5min. Electrophoresis was carried out at 80V for 3h in an agarose gel at 2.5%, dyed with SYBR Safe™ DNA Gel Stain 1×. For the visualization and analysis of the fragment sizes obtained, the Gel Doc EZ™ Bio-Rad image documenter was used.
ResultsPhenotypical identification of B. abortus isolatesThe microscopic morphology using Gram stain showed gram-negative coccobacilli, and by using Stamp's staining, red dyed coccobacilli were observed because the genus Brucella has the property of being partially acid-alcohol resistant. All isolates showed agglutination with the anti-Brucella abortus polyclonal serum (Difco). Biochemical tests showed that all isolates were positive to oxidase and catalase with intermediate urease enzyme activity (the reaction occurs approximately after 2h of incubation). All isolates produced H2S on strips of lead acetate and after several passes the isolates did not need CO2 for their growth with the exception of isolates Ba-Col-B-006 and Ba-Col-B-012 isolates. Likewise, all the isolates grew in the presence of different concentrations of basic fuchsine or safranin; however, they did not grow in the presence of thionine.
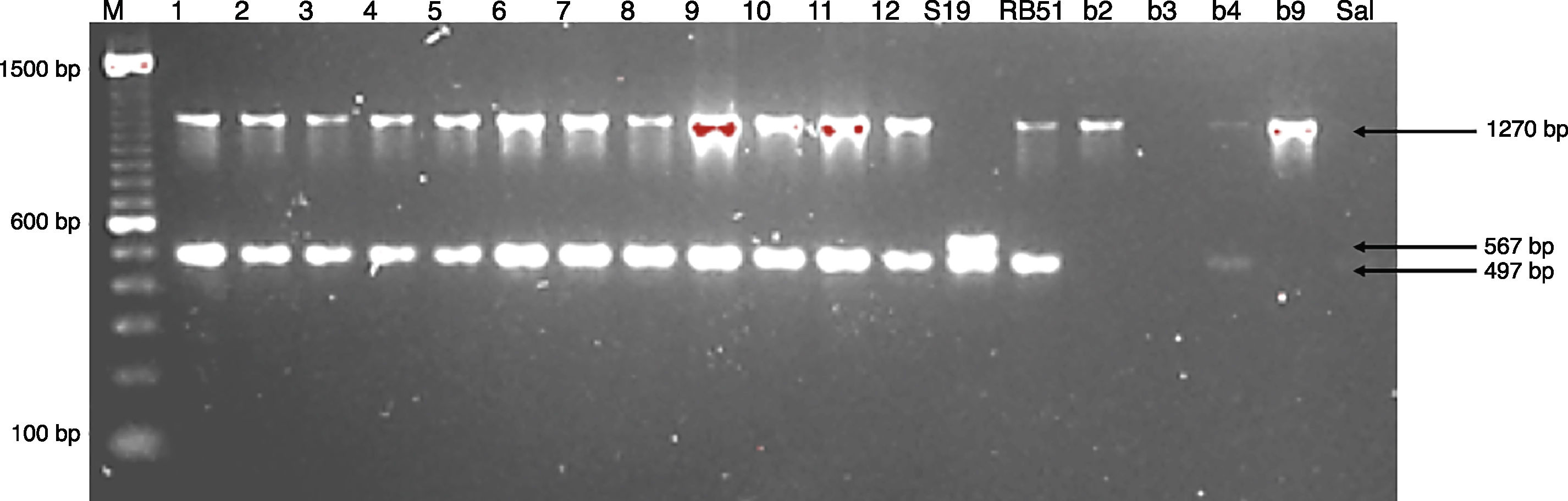
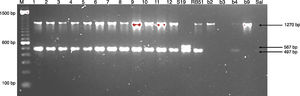
Molecular identificationAMOS-ERY-PCRWith the multiple PCR using AMOS-ERY primers, a 1270bp fragment band was observed, corresponding to a region of the eryC–eryD genes in charge of erythritol catabolism in all members of the Brucella genus. Furthermore, in the twelve isolates and in the two vaccine strains, the amplification of a species-specific B. abortus fragment of approximately 497bp was observed (Fig. 1).
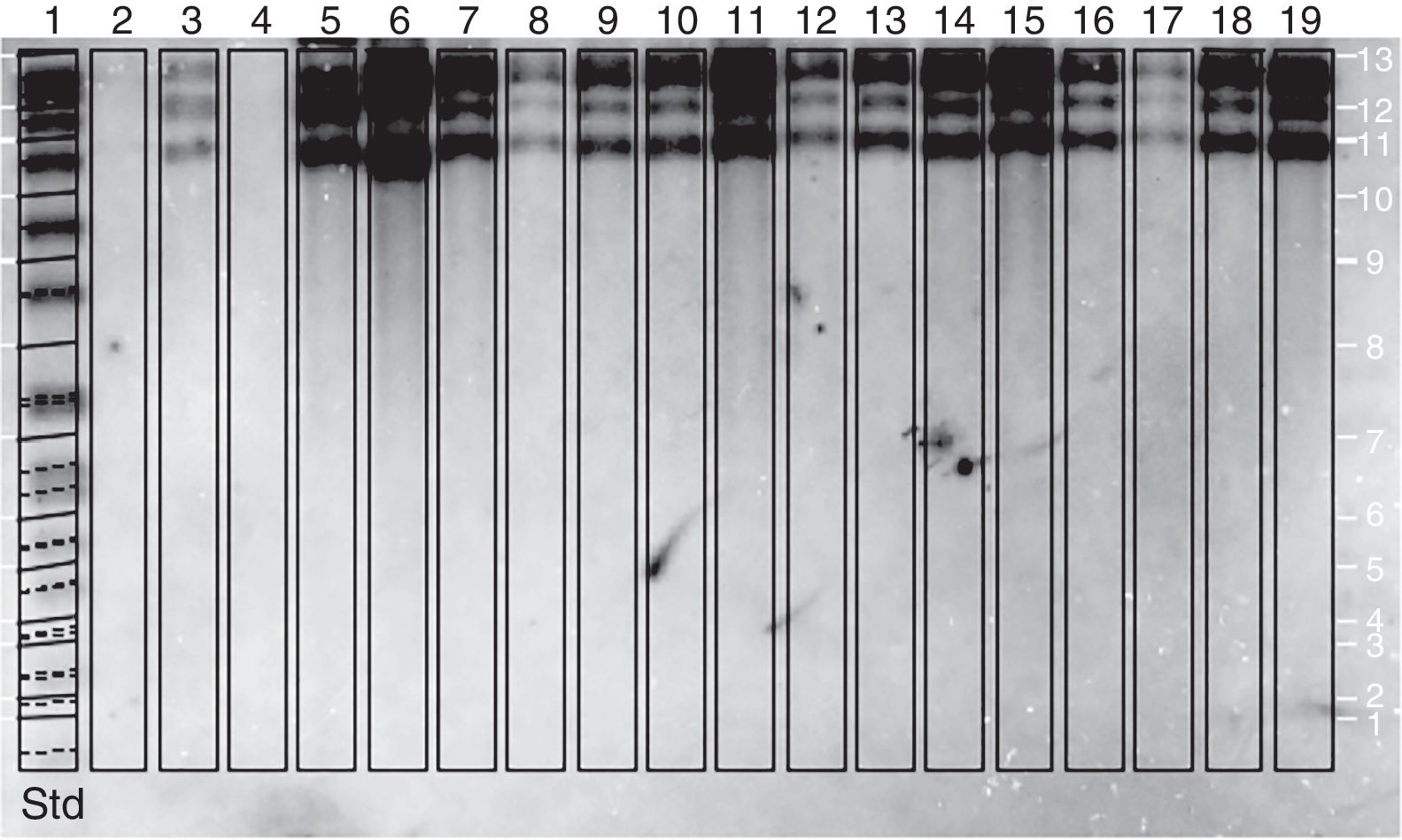
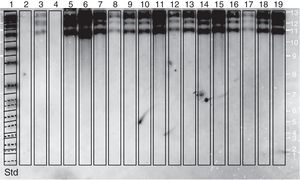
Restriction fragment length polymorphism analysis–IS711 (RFLP–IS711) and detection by Southern blotAn RFLP-IS711 analysis was carried out to try to differentiate biovars and genetic variants among the twelve isolates used in this study. This technique generated different characteristic hybridization patterns. The hybridization band sizes ranged approximately between 4.8 and 9.0kbp. Four copies of IS711 were found in each pattern (Fig. 2).
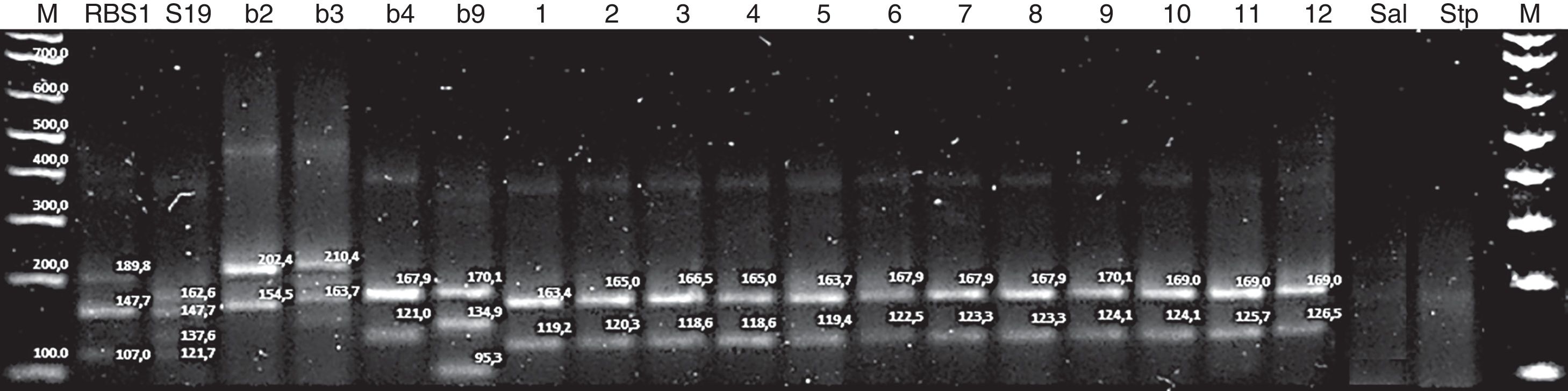
Multiple locus variable number of tandem repeat analysis (MLVA) using multiple PCRBased on two selected VNTR loci, the multiple PCR called MLVA-2 allowed to easily obtain differentiated genetic patters in agarose gels, both for vaccine RB51 strains and for B. abortus strain 19, as well as for the four reference biovars (B. abortus bv. 2, B. abortus bv. 3, B. abortus bv. 4 and B. abortus bv. 9). In the multiple PCR (Fig. 3) the twelve isolates showed the same MLVA pattern that is also similar to B. abortus bv. 4 reference biovar, whose fragment is approximately equivalent to 168bp (locus 1) and to 124bp (locus 7). The homologous fragments in the isolates ranged from 163 to 170bp (locus 1) and from 119 to 126bp (locus 7).
Multiple PCR-MLVA-2 for RB51 (Vaccine strain RB51, biovar 1), S19 (vaccine strain S19, biovar 1), b2 (reference strain B. abortus, biovar 2), b3 (reference strain B. abortus, biovar 3), b4 (reference strain B. abortus, biovar 4), b9 (reference strain B. abortus, biovar 9), 1–12 isolates of B. abortus, Sal: Salmonella enteritidis and Stp: Staphylococcus aureus; M: DNA marker 100bp Invitrogen®.
Based on the morphology observed under the Gram and Stamp stains and the phenotypic test results, it was confirmed that the isolates used in this study belong to the B. abortus species. The results obtained with conventional phenotyping methods such as the test of sensitivity to thionine and resistance to basic fuchsine and safranin suggest that all the isolates belong to biovar 1 or 4. All isolates and the RB51 vaccine strain grew in the presence of erythritol in contrast to vaccine strain 19, confirming that the isolates are field strains and not of vaccine origin12,40,43.
For the AMOS-ERY PCR amplification a fragment of 1270bp was observed in all the twelve isolates and in the RB51 vaccine strain but not in the S19 vaccine, in which a fragment of 567bp was observed due to the deletion of 702bp in the eryC–eryD genes. This finding confirms that the genus of the twelve isolates was Brucella. Moreover, it was possible to differentiate the twelve field isolates from strain 19 vaccine.
Through this amplification it was possible to obtain a 497bp fragment in the twelve isolates and in the vaccine strains. Since this fragment is common in B. abortus biovars 1, 2 and 4, this methodology cannot be considered for the discrimination among this three biovars2,8,12,30,33. The patterns obtained with the RFLP-IS711 method coincide with the findings reported by Bricker and Halling8 and Ocampo et al.30 in which no differentiation among these three biovars was observed. The RFLP-IS711 using the Southern blot technique only identified a profile but no genetic variability was observed within the twelve isolates. This suggests that these isolates belong to the same biovar or that this technique does not have the sufficient resolution power to discriminate among the three biovars of B. abortus11.
The genetic homogeneity observed in the twelve isolates indicates the need to complement this molecular identification at the biovar level using other molecular techniques focusing on genetic markers with a much higher resolution power to subtype this microorganism.
The results obtained with the MLVA technique showed small size differences (approximately 7bp) in the fragments of locus 1 and locus 7, which might be due to deletions or insertions in these hypervariable VNTRs, causing an increase or decrease in the number of repeated units in each locus. Therefore, these differences can be observed with differences in the size of the bands in the agarose gel. This might indicate that although it is the same biovar, there are allelic variants within itself, and that the VNTR resolution power to identify genetic variants in the biovars of the MLVA scheme was useful. The MLVA scheme shows minimum genetic loci differences besides the genetic homogeneity that the members of the genus Brucella have5,1,29.
The multiple PCR also shows that the two vaccine strains and the reference biovars 3 and 9 show several bands. This might be due to the presence of multiple alleles in any of the two loci that were used. Moreover, it can also be observed that although the two vaccine strains are classified as biotype 1, they show a different genetic pattern, which coincides with the results obtained by Bricker et al.6 and Whatmore et al.41 These authors report the same event with vaccines and reference strains that have been subcultivated for a long time.
Due to the high variability of the loci chosen for the MLVA-2 scheme, this technique becomes a complementary methodology for reliable taxonomical identification. However, as the MLVA-2 scheme proposed in this work is a multiple PCR, it can be considered a quick and economic technique to identify isolates, having the additional advantage of being able to easily visualize biovar differential patterns. Tracking of this HOOF-print stability strategy should be developed in strains and field isolates maintained under laboratory conditions, as well as in loci VNTR in order to evaluate the hypervaribility changes of the two genetic markers reported in this study.
It has been observed that B. abortus biovars have specific geographic distributions, e.g. B. abortus biovars 1 and 2 can be found almost everywhere around the world. Biovars 3 and 4 are less frequent, although biovar 4 has been reported in Argentina, Brazil, Chile, Cuba, Ecuador, Mexico, and Venezuela15,21,27. In Colombia, most of the studies have focused on the seroprevalence of B. abortus in the country in animals as well as in humans9,20. However, the few studies that have focused on the identification of B. abortus strains that circulate in Colombia report the presence of B. suis bv. 1, B. canis and B. abortus bv. 1, 2, and 4, being B. abortus bv. 1, the most commonly found and associated with diseased animals. It must be highlighted that similar studies aiming to genotype Brucella spp. isolates have been carried out in international reference centers in countries such as Argentina by using conventional and phagotyping methods10,21.
The identification in this study of twelve strains of B. abortus biovar 4 is consistent with the work by Pacheco et al.31 in which the analysis of the complete genome of the isolation Ba-Col – B-012 allowed to identify it as B. abortus biovar 4. A complementary identification will be done by serological classification with monospecific anti-A, anti-M, which agglutinate biovar 1 and biovar 4, respectively. This will have to be conducted in a Brucella Reference Laboratory. The identification of the twelve isolates as B. abortus biovar 4 cannot be considered a rare event since these isolates were obtained in the department of Nariño13 and according to studies carried out by Lucero et al.21 with isolates acquired between 1968 and 1991 in 15 Latin American countries, this biovar was reported in Colombia as well as in Ecuador. Recent studies conducted by Ron-Román et al.34 and Rodríguez et al.33 in northeast Ecuador in the border with Colombia, the presence of B. abortus bv. 4 has been reported in humans as well as in animals. Similarly, Minharro et al.27 identified this biovar in Rio Grande do Sul, Brazil. In this regard, it will be necessary to check the origin of these biovars, and if there is a relationship between B. abortus bv. 4 isolates in Colombian cattle with other isolates from the region, due to the transportation of infected animals from other regions or trade of semen or other biological material that could lead to biovar spreading.
Research works that define the presence of different biovars in Colombia as well as in other South American countries can be of great help to understand not only the epidemiological dynamics of the disease, but also to support the surveillance and control program of bovine brucellosis in the country as well as in animals and humans. Finally, this information will help to establish studies to carry out the epidemiological tracking of groups or clonal complexes of different biovars in the continent.
ConclusionsThis study allowed the identification of Colombian bovine B. abortus isolates to biovar level by using conventional and molecular techniques with enough resolution power. Although several studies report B. abortus biovars 1 and 2 as the most common biovars found in Colombia, the phenotypic and molecular identification carried out in this study confirmed that the twelve Colombian isolates obtained from dairy farms in Nariño, Colombia during the 1990s belonged to B. abortus biovar 4. This is first study to focus on the phenotypic and molecular identification of B. abortus isolates in Colombia. However, due to the fact that this study was limited to a small geographical area, other studies should be conducted at the national level to identify the presence of biovars that could be present in the country. Moreover, further information regarding their distribution is also needed to contribute to a better comprehension of bovine brucellosis in Colombia.
Conflict of interestThe authors declare that they have no conflicts of interest.
The authors want to thank the Ministry of Agriculture and Rural Development of Colombia (MADR) for funding this study through Instituto Colombiano Agropecuario (ICA) that we are also very grateful to for trusting us to implement this study via AGROSAVIA's Microorganism Germplasm Bank of Interest in Animal Health-Bacteria and Viruses (BGSA-BV). Moreover, we want to express our gratitude to AGROSAVIA for providing all the infrastructure necessary to conduct the entire research and for managing all the administrative issues that imply a large workload.