In this study, the culture analysis of urine samples from patients hospitalized in the Central-West region of Brazil was performed, and the isolated microorganisms were phylogenetically identified as Trichosporon asahii. Maximum parsimony analysis of the IGS1 sequences revealed three novel genotypes that have not been described. The minimum inhibitory concentrations of the nine isolates identified were in the range of 0.06–1μg/ml for amphotericin B, 0.25–4μg/ml for fluconazole, and 0.03–0.06μg/ml for itraconazole. Approximately 6/9 of the T. asahii isolates could form biofilms on the surface of polystyrene microplates. This study reports that the microorganisms isolated here as T. asahii are agents of nosocomial urinary tract infections. Furthermore, the IGS1 region can be considered a new epidemiological tool for genotyping T. asahii isolates. The least common genotypes reported in this study can be related to local epidemiological trends.
En este estudio fueron analizadas mediante el cultivo muestras de orina de pacientes hospitalizados en la región centro-oeste de Brasil; los microorganismos aislados fueron identificados filogenéticamente como Trichosporon asahii. A través del análisis de máxima parsimonia de las secuencias de IGS1, fueron encontrados 3 genotipos que no habían sido descritos anteriormente. Las concentraciones inhibitorias mínimas frente a los 9 aislados identificados presentaron un rango de 0,06-1μg/ml en el caso de la anfotericina B, de 0,25-4μg/ml en el del fluconazol, y de 0,03-0,06μg/ml en el del itraconazol. Aproximadamente 6/9 de los aislados de T. asahii formaron biopelículas en la superficie de microplacas de poliestireno. Este trabajo documenta el aislamiento de T. asahii como agente causal de infeciones urinarias nosocomiales. Además, demuestra que la región IGS1 puede ser considerada una nueva herramienta epidemiológica para la genotipificación de los aislados de T. asahii. Los genotipos menos comunes encontrados en este estudio pueden estar relacionados con las características epidemiológicas locales.
Trichosporon causes superficial and systemic nosocomial infections2,11. These yeasts have been found to be the causative pathogens of white piedra, and opportunistic pathogens in invasive infections in immunocompromised patients in tertiary hospitals worldwide2. However, the urinary tract infection caused by Trichosporon asahii is the most prevalent, regardless of the geography6,7,16.
The objective of this study was to investigate the genotypes, antifungal susceptibility, and biofilm formation of T. asahii isolates obtained from urine of patients admitted to the University Hospital, Federal University of Grande Dourados, Dourados, Central-West Brazil.
Between October 2010 and January 2011, 18 patients admitted to this hospital were diagnosed with urinary tract infection (mainly fungal on the basis of clinical and laboratory findings). Nine (9) pure T. asahii isolates with >105 colony-forming units per millilitre were obtained from 6 of these 18 patients (33%). These isolates were analyzed phenotypically and confirmed by using molecular techniques. A remarkable observation is the large number of urinary tract infection cases that were identified over the span of four months.
The purity of the cultures was confirmed by growth on CHROM agar Candida® (Difco®, Sparks, MD, USA) and yeasts were identified in accordance with the classical methodology based on macroscopic, microscopic, and biochemical characteristics9.
DNA sequencing was performed to confirm identification: Trichosporon DNA was extracted using the method proposed by Chong et al.3. To accurately identify the Trichosporon strains to species level, the IGS1 region was amplified by PCR (ABI 3730 XL; Applied Biosystems, Foster City, CA) and purified using the ABI PRISM Big Dye Terminator Cycle Sequencing Kit, according to the manufacturer's instructions. The primers used were 26SF (5′-ATCCTTTGCAGACGACTTGA-3′) and 5SR (5′-AGCTTGACTTCGCAGATCGG-3′), and sequencing was performed according to Sugita et al.’s method14.
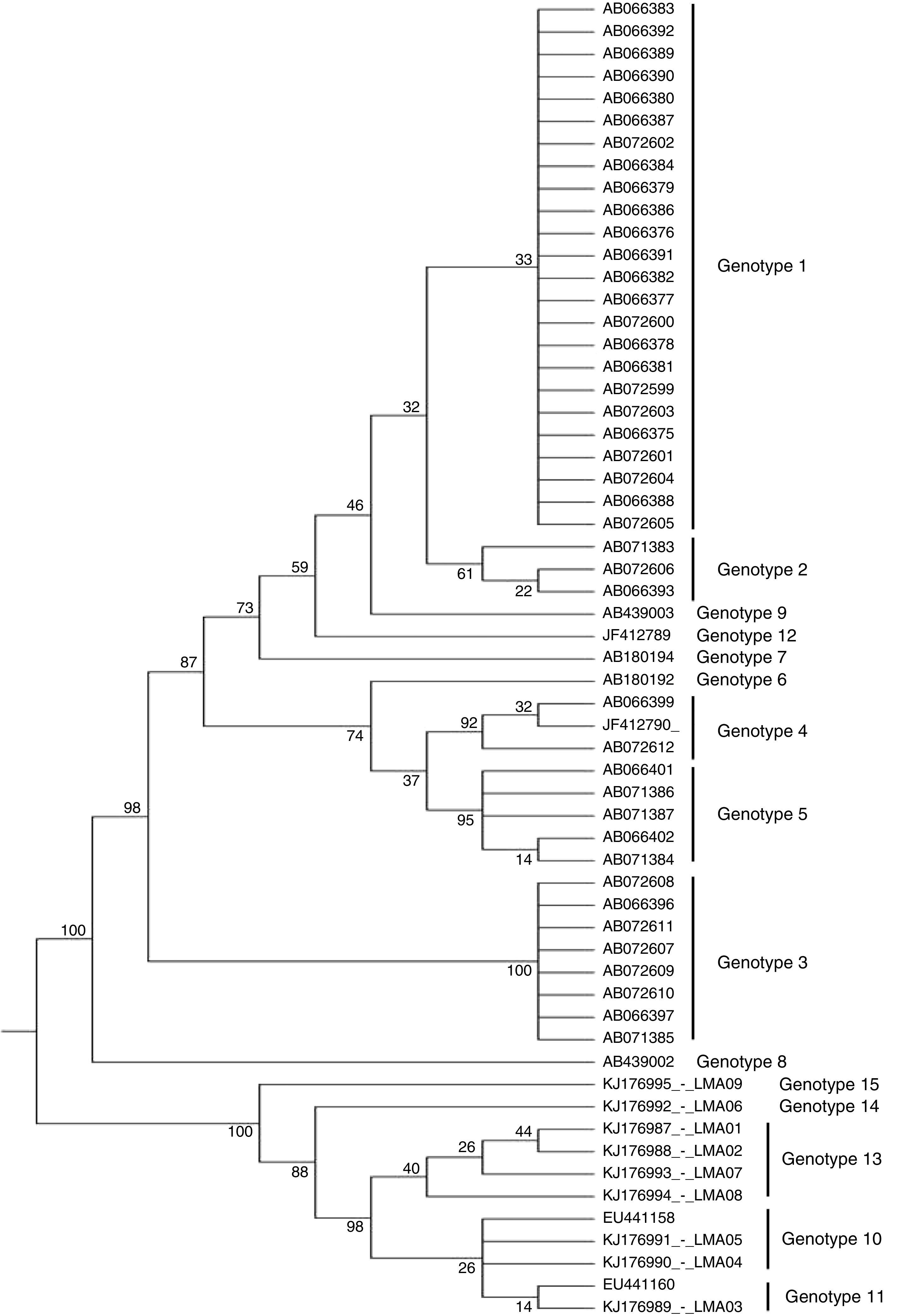
Molecular epidemiology markers were assessed by T. asahii genotyping and phylogenetic analysis. The IGS1 sequences of the T. asahii strains determined in this study were compared with DNA sequences deposited in GenBank. All phylogenetic analyses were performed using the Mega 6 software. The phylogenetic tree was constructed using the unweighted pair group method with average linkages2. The phylogram stability was assessed by parsimony bootstrapping with 1000 simulations.
The antifungal susceptibility profile was determined by broth microdilution, in accordance with the M27-A3 document of the Clinical and Laboratory Standards Institute4, testing the following antifungals: amphotericin B (Bristol – Myers Squibb), itraconazole (Janssen), and fluconazole (Pfizer).
Biofilm production was evaluated on the surfaces of polystyrene microplates using the method described by Shin et al.12.
T. asahii seems to be the most common species isolated from the urine of hospitalized patients in Brazil5,13, followed by Trichosporon inkin and Trichosporon asteroides. In addition, T. asahii is the most common species identified in disseminated trichosporonosis5.
The nine isolates were phylogenetically identified as T. asahii and the maximum parsimony analysis of the IGS1 sequence revealed three novel genotypes that have not been yet described in Brazil and worldwide (Fig. 1), which were named as genotype 13 (GenBank accession numbers: KJ176987, KJ176988, KJ176993, KJ176994), 14 (accession number: KJ176992), and 15 (accession number: KJ176995). The other three were identified as genotype 10 (accession numbers: KJ176991 and KJ176990) and 11 (accession number: KJ176989).
Molecular phylogenetic tree of T. asahii strains based on confidently aligned IGS1 sequences, including those from reference strains and the novel genotype strains found in Brazil. The code numbers correspond to the GenBank accession numbers. The values were obtained using maximum parsimony cluster analysis and 1000 bootstrap simulations.
Interestingly, genotyping showed mixed infections by two T. asahii genotypes: isolates 2 (genotype 13) and 4 and 5 (genotype 10) were recovered from the same patient and, similarly, isolates 6 (genotype 14) and 9 (genotype 15) were recovered from another patient. However, this phenomenon was reported earlier16.
Currently, there are 12 known genotypes of T. asahii17, however, in Brazil, the vast majority (>80%) of the isolates genotyped so far belongs to genotype 1, followed by genotypes 3, 4, and 62,5. Our findings may indicate the identification of novel genotypes that are not so common in Brazilian hospitals. In fact, further studies need to be conducted considering the characteristics of each hospital and the geography of Brazil. To the best of our knowledge, this is the first documentation of the molecular characterization of Trichosporon isolates identified in the Brazilian countryside.
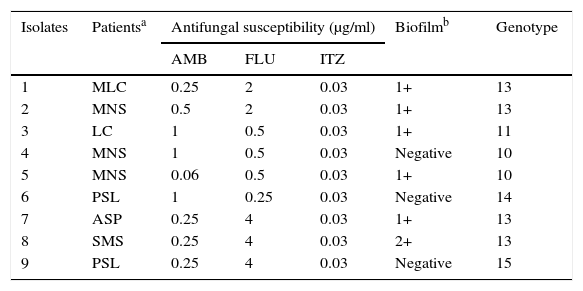
The minimal inhibitory concentration (MIC) of the nine isolates was in the range of 0.06–1μg/ml for amphotericin B, 0.25–4μg/ml for fluconazole, and 0.03–0.06μg/ml for itraconazole (Table 1). There are no defined criteria to determine the cutoff point in order to establish the susceptibility profile of Trichosporon. Interestingly, the MIC was low for all of our isolates for the antifungal agents tested, contradicting other studies on T. asahii susceptibility to triazoles and amphotericin B8,10.
Results from the nine isolates of T. asahii
| Isolates | Patientsa | Antifungal susceptibility (μg/ml) | Biofilmb | Genotype | ||
|---|---|---|---|---|---|---|
| AMB | FLU | ITZ | ||||
| 1 | MLC | 0.25 | 2 | 0.03 | 1+ | 13 |
| 2 | MNS | 0.5 | 2 | 0.03 | 1+ | 13 |
| 3 | LC | 1 | 0.5 | 0.03 | 1+ | 11 |
| 4 | MNS | 1 | 0.5 | 0.03 | Negative | 10 |
| 5 | MNS | 0.06 | 0.5 | 0.03 | 1+ | 10 |
| 6 | PSL | 1 | 0.25 | 0.03 | Negative | 14 |
| 7 | ASP | 0.25 | 4 | 0.03 | 1+ | 13 |
| 8 | SMS | 0.25 | 4 | 0.03 | 2+ | 13 |
| 9 | PSL | 0.25 | 4 | 0.03 | Negative | 15 |
AMB, Amphotericin B; FLU, Fluconazole; ITZ, Itraconazole.
The adherent biofilm layer was scored as either negative or weakly (1+), moderately (2+ or 3+), or strongly (4+) positive12.
The ability to form biofilm on the surfaces of polystyrene microplates was observed in 6/9 of the isolates of T. asahii (Table 1), though the degree of biofilm formation was small: 1+ (55.5%) and 2+ (11.1%). Trichosporon spp. are known to form biofilms1,15.
In conclusion, this study reports that the T. asahii microorganisms isolated here are agents of nosocomial urinary tract infections. This is the first study in the Central-West region of Brazil that identified novel genotypes not been previously described. Moreover, different genotypes were isolated from the same patient. The IGS1 region may be an epidemiological tool for genotyping T. asahii isolates. The least common genotypes reported in our study can be related to local epidemiological trends.
Ethical disclosuresProtection of human and animal subjectsThe authors declare that the procedures followed were in accordance with the regulations of the responsible Clinical Research Ethics Committee and in accordance with those of the World Medical Association and the Helsinki Declaration.
Confidentiality of dataThe authors declare that no patient data appears in this article.
Right to privacy and informed consentThe authors declare that no patient data appears in this article.
Conflict of interestThe authors declare that they have no conflicts of interest.
This work was supported by Fundação de Apoio ao Desenvolvimento do Ensino, Ciência e Tecnologia do Estado de Mato Grosso do Sul (FUNDECT) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).