To describe a clonal outbreak due to vancomycin-resistant Enterococcus faecium (VREF) in the nephrology and renal transplant unit of a tertiary teaching hospital in Barcelona, Spain, and to highlight how active patient and environment surveillance cultures, as well as prompt and directed intervention strategies, mainly environmental, helped to successfully bring it under control.
Patients and methodsA study was conducted on patients admitted to the nephrology ward with any culture positive for VREF over a 6-month period (August 2012–January 2013). Based on the identification of a clonal link between the isolates, weekly rectal screening using swabs was implemented for all patients, as well as environmental cultures and cleaning of medical equipment and the ward. VREF isolates were identified by MicroScan and confirmed by Etest. Bacterial identification was confirmed by MALDI-TOF MS. The presence of van genes, and esp and hyl virulence genes was determined using PCR. The clonal relationship between the isolates was studied first with DiversiLab (bioMérieux), and then by PFGE-Smal and MLST. A two-tier sequence of infection control measures was implemented.
ResultsDuring the study period, VREF was isolated from 13 patients. All cases were colonized with no criteria for infection. VREF isolates were also extensively recovered from the environment and medical equipment. Isolates carried the vanA gene, and were multidrug-resistant, including high-level resistance (MIC >16mg/L) to vancomycin and teicoplanin. Molecular analysis showed that all VREF isolates belonged to sequence type 17 (ST17) carrying hyl virulence genes. After implementing infection control measures in a two-tier sequence, and reinforcing particularly environmental and medical equipment cleaning, no further cases were detected in the follow-up year.
ConclusionA clonal outbreak of VREF-ST17 involving only colonization is reported. The prompt implementation of aggressive infection control measures in patients and the environment was effective in controlling the outbreak and avoided the potential emergence of infection among patients.
Describir un brote clonal de Enterococcus faecium resistente a vancomicina (VREF) en la unidad de trasplante renal de un hospital universitario en Barcelona (España) y destacar que los controles ambientales, así como las estrategias de intervención dirigidas y tempranas, principalmente ambientales, fueron suficientes para controlar el brote.
Pacientes y métodosSe estudiaron todos los pacientes ingresados en la unidad de nefrología con un cultivo positivo para VREF en un periodo de 6 meses (Agosto de 2012 a Enero de 2013). Basados en la identificación de una relación clonal entre las cepas, se implementaron frotis rectales de cribado para todos los pacientes, así como frotis ambientales y limpieza de todo el material médico y de la unidad. Se identificaron las cepas de VREF por MicroScan y se confirmaron con Etest. La identificación bacteriana se confirmó con MALDI-TOF MS. La presencia de genes van, y de genes de virulencia esp y hyl, se investigó por PCR. La relación clonal entre las cepas se estudió con DiversiLab (bioMérieux), y después con PFGE-Smal y MLST.
ResultadosDurante el periodo estudiado, se aislaron cepas de VREF de 13 pacientes. Todos fueron casos de colonización sin casos de infección. Se aislaron numerosas cepas del ambiente y del equipo médico. Las cepas presentaban el gen VanA y eran multirresistentes. El análisis molecular mostró que todas las cepas pertenecían a la secuencia tipo17 (ST17), portando genes de virulencia hyl. Tras la implementación de medidas de control de infección de 2 niveles, e incrementando sobretodo la limpieza del ambiente y del equipo médico, no se detectaron nuevos casos en el año posterior.
ConclusiónSe informa de un brote clonal de VREF-ST17. La pronta implementación de medidas agresivas de control de infección en pacientes y en el ambiente fue efectiva para el control del brote.
The first isolates of vancomycin-resistant Enterococcus faecium (VREF) were detected in England and France in 1986. Since then, it has become an increasingly common pathogen in nosocomial infections, especially among immunosuppressed patients. VREF can cause a range of infections, including bacteremia, urinary tract infections and intraabdominal infections, among others.1–3 In the United States, colonization, leading to infection in some patients, is endemic in many hospitals and has been related to antibiotic use. In 1989 the CDC recognized E. faecium as the second most common pathogen in nosocomial infections,4,5 the result in fact of the transmission capacity of the bacteria, with immunocompromised patients acting as hosts.6 In Europe, VREF exists in the form of outbreaks, and hence epidemics, whose origin is believed to be related to the use of avoparcin, a vancomycin-like glycopeptide widely used in agriculture.7 The nosocomial transmission of VREF lies in its ability to persist on environmental surfaces and medical devices, as well as to survive high temperatures, heat and some alcohol preparations.8 The risk factors for acquisition of VREF infection include contact with patients colonized or infected with VREF, previous administration of antibiotics, long-term hospitalization (especially in intensive care or surgical facilities) and solid organ and bone marrow transplants, among others.9 Virulence factors include gelatinase production, surface protein (esp) involved in biofilm formation, and the aggregation substance (AS).10 Even though VREF can be found in healthy individuals, the genetic features and virulence factors such as the esp gene have been found during some outbreaks.11
Spain has one of the lowest rates of VREF in Europe, being mainly limited to sporadic outbreaks with the presence of VREF carrying the vanA gene. The vanB phenotype is less prevalent and was first described in 2001.12 In the rest of Europe, the prevalence is higher, with vanA VRE being most prevalent in the UK (2.7%) and vanB VRE in Slovenia (2%).13
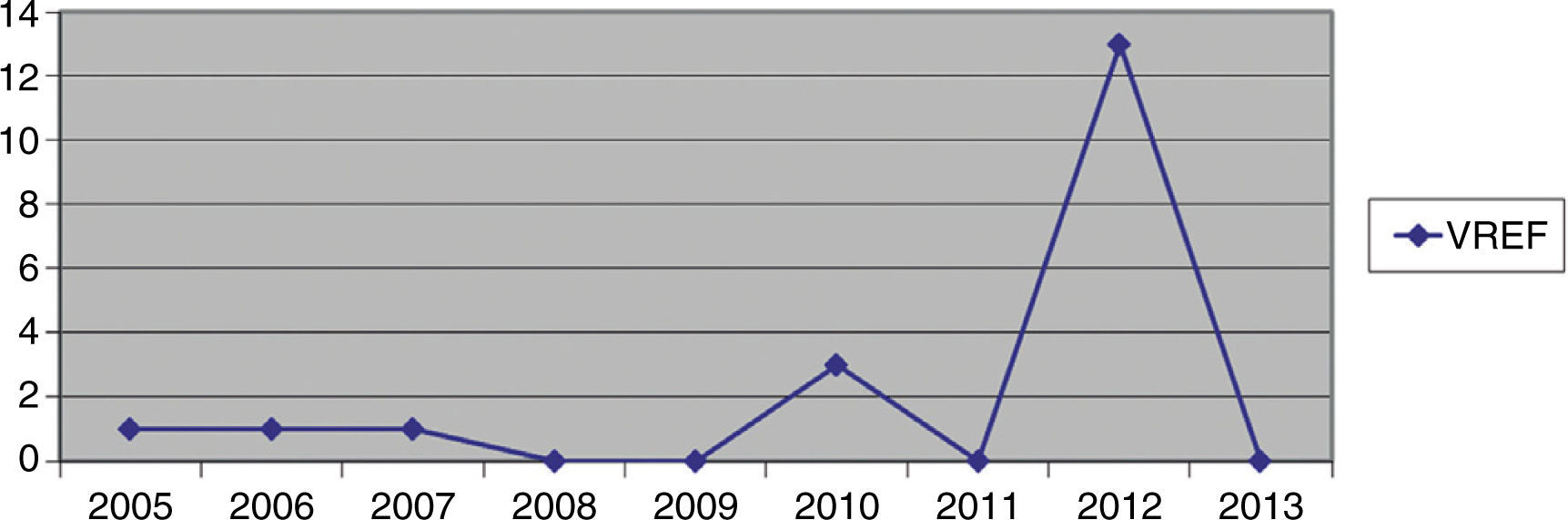
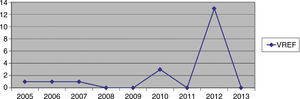
Hospital Del Mar is a university hospital in Barcelona, Spain. Sporadic cases of VREF have been reported every year between 2005 and 2012, with a maximum of 3 cases per year (Fig. 1). Thirteen VREF isolates were detected in the nephrology ward of our hospital between August 2012 and January 2013. Our objective is to report a clonal outbreak of VREF in the nephrology and renal transplant ward of a tertiary teaching hospital in Barcelona, Spain, which we believe is important to share in this era of growing multiresistance; and to highlight how active surveillance cultures of patients and the environment, as well as prompt and directed infection control strategies, mainly environmental, helped control the outbreak rapidly and successfully.
MethodsOrion statement guidelines for transparent reporting of outbreak reports were followed.14 This study was done retrospectively.
Setting and patient populationThe Hospital Del Mar is a 400-bed acute care university hospital in Barcelona, Spain. The nephrology unit has a total of 14 beds, with bed occupancy of 4665 inpatient days in 2012 (92%). From August 2012 through June 2013, epidemiological and clinical data of the involved patients were collected.
Case definitionA case of VREF colonization or infection was established as any patient admitted to hospital with a confirmed infection or colonized with an isolate of vancomycin and ampicillin-resistant E. faecium in the 48h following admission.
Surveillance cultures and VREF detectionFrom November 2012 through June 2013, a rectal screening with cotton swabs was collected from every patient admitted to the nephrology ward and this practice was repeated weekly. Previous to the outbreak patients were routinely screened for multiresistant bacterial colonization as they are admitted to wards with immunosuppressed patients. Environmental surveillance cultures were collected from every room to which a patient with a positive rectal swab had been admitted from November 2012 to January 2013. These cultures were performed using moistened sterile gauze pads.
After this date, rectal swabs were performed at admission only. Rectal swabs were cultured in Campylobacter on blood agar plates (BioMerièux) and incubated at 37°C for 24–48h.
Colonies grown from surveillance and environmental samples were selected for further microbiological studies. Bacterial identification was confirmed by the MALDI-TOF MS method (Bruker Daltonics, Germany). Susceptibility testing for ampicillin, erythromycin, tetracycline, teicoplanin, vancomycin, linezolid and high-level gentamicin and streptomycin resistance was performed by microdilution, using the automated MicroScan System (Beckman Coulter, CA) and interpreted according to CLSI criteria (ref: Clinical Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. Nineteenth Informational Supplement M100-S25. Wayne, PA, USA: CLSI, 2015). Resistance to vancomycin and teicoplanin was confirmed by Etest (BioMérieux). The presence of van genes was investigated by PCR.15,16
Clonal relatednessClonal relationships among the isolates were first studied with the commercial rep-PCR fingerprinting system (DiversiLab System, BioMerieux)), using the Pearson correlation coefficient. Isolates that clustered at ≥95% were considered as related. Clonal relationships among isolates were also established by PFGE-SmaI and MLST (http://efaecium.mlst.net/)
Enterococcal virulence genesThe presence of esp (enterococcal surface protein) and hyl (glycosyl hydrolase) genes was investigated by PCR.17
Infection control measuresBefore November 2012, patients carrying or infected by VREF were managed according to Centers for Disease Control and Prevention (CDC) Tier 1 precautions,17 namely, isolation in individual rooms, contact precautions including gloves, gowns, and hand hygiene, and daily room cleaning with sodium hypochlorite 1000ppm (Sprint H-100®) or a combination of quaternary ammonium and amines (Sprint H-200®) on surfaces where sodium hypochlorite would be corrosive. After November 2012, CDC Tier 2 measures were applied to all patients admitted to the unit17 with isolation in individual rooms independent of carrier status. As part of CDC Tier 2 measures, room cleaning was reinforced, being performed more frequently and extensively, paying special attention to inanimate surfaces; the concentration of sodium hypochlorite 1000ppm (Sprint H-100®) was increased to 5000ppm. Cleaning was monitored by means of weekly environmental surveillance swabs, which included surfaces in close proximity to the patient and staff. The number of cleaning staff was increased at this point. All medical equipment associated with individual patients was cleaned after use. Staff changed their uniforms daily. Weekly meetings were scheduled with the ward staff and infection control team in order to inform, update and reinforce measures. Members of staff were retrained to ensure correct hand hygiene and the cleaning staff was also retrained. Patients and families were informed of the outbreak and given instructions on isolation and cleaning measures. Only one person was allowed to visit the patient at any one time. New admissions were not allowed in the ward. Ward doors were closed to minimize the number of people entering the unit. No changes in antibiotic policy were made at his point.
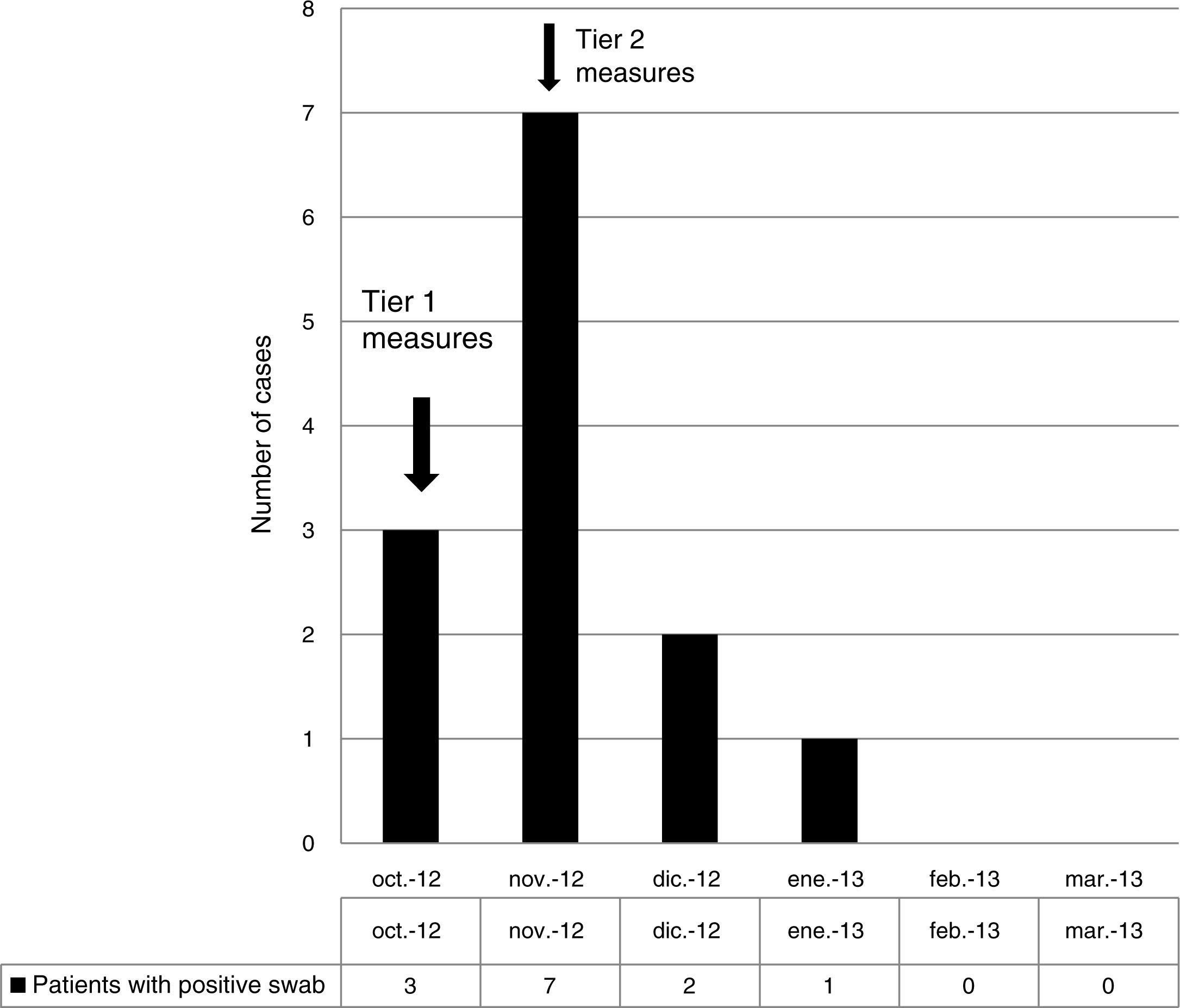
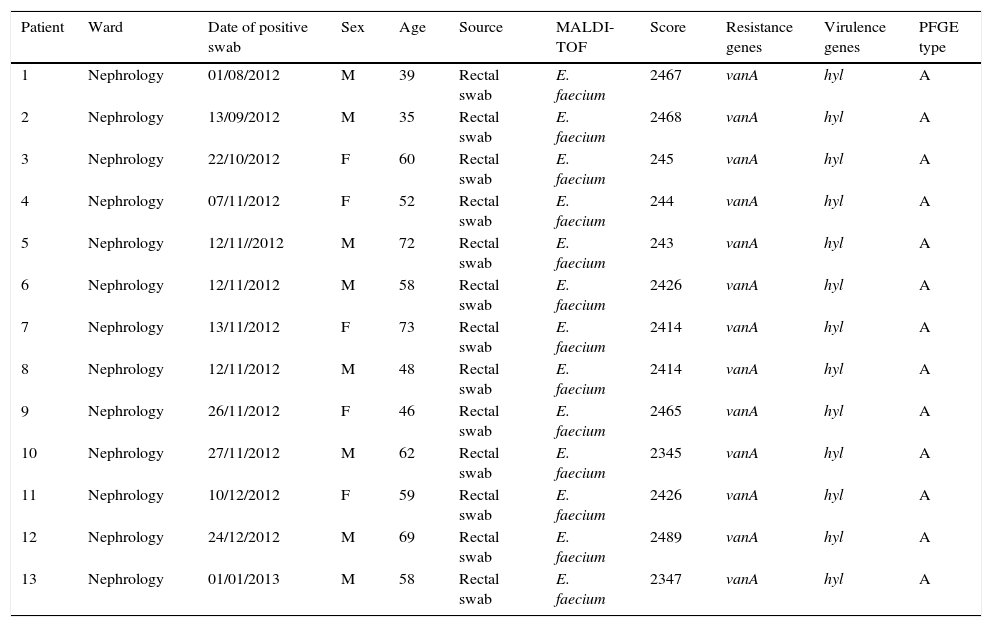
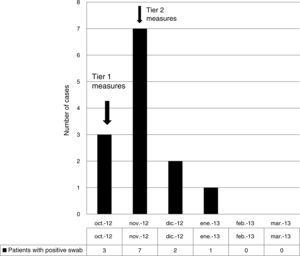
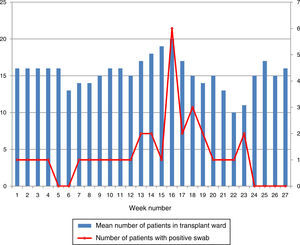
ResultsOutbreak description and interventionBetween 0 and 3 cases of VREF per year are normally reported at our hospital (Fig. 1). Patients are routinely screened for multiresistant bacterial colonization as they are admitted to wards with immunosuppressed patients. In August and September 2012, two patients were reported as colonized by VREF in the nephrology and renal transplant unit. In October 2012, a third case with VREF fecal colonization was reported in the same unit. After that, an outbreak was declared due to the rapid increase in the number of cases (Fig. 2). During the outbreak, the incidence density of VREF nosocomial acquisition in the nephrology ward was 9.23 per 1000 patient days, compared with 0 in the same period in 2011 (p<0.001). The last case reported in the nephrology ward was in January 2013. Characteristics of patients are illustrated in Table 1.
Characteristics of patients and isolates.
| Patient | Ward | Date of positive swab | Sex | Age | Source | MALDI-TOF | Score | Resistance genes | Virulence genes | PFGE type |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Nephrology | 01/08/2012 | M | 39 | Rectal swab | E. faecium | 2467 | vanA | hyl | A |
| 2 | Nephrology | 13/09/2012 | M | 35 | Rectal swab | E. faecium | 2468 | vanA | hyl | A |
| 3 | Nephrology | 22/10/2012 | F | 60 | Rectal swab | E. faecium | 245 | vanA | hyl | A |
| 4 | Nephrology | 07/11/2012 | F | 52 | Rectal swab | E. faecium | 244 | vanA | hyl | A |
| 5 | Nephrology | 12/11//2012 | M | 72 | Rectal swab | E. faecium | 243 | vanA | hyl | A |
| 6 | Nephrology | 12/11/2012 | M | 58 | Rectal swab | E. faecium | 2426 | vanA | hyl | A |
| 7 | Nephrology | 13/11/2012 | F | 73 | Rectal swab | E. faecium | 2414 | vanA | hyl | A |
| 8 | Nephrology | 12/11/2012 | M | 48 | Rectal swab | E. faecium | 2414 | vanA | hyl | A |
| 9 | Nephrology | 26/11/2012 | F | 46 | Rectal swab | E. faecium | 2465 | vanA | hyl | A |
| 10 | Nephrology | 27/11/2012 | M | 62 | Rectal swab | E. faecium | 2345 | vanA | hyl | A |
| 11 | Nephrology | 10/12/2012 | F | 59 | Rectal swab | E. faecium | 2426 | vanA | hyl | A |
| 12 | Nephrology | 24/12/2012 | M | 69 | Rectal swab | E. faecium | 2489 | vanA | hyl | A |
| 13 | Nephrology | 01/01/2013 | M | 58 | Rectal swab | E. faecium | 2347 | vanA | hyl | A |
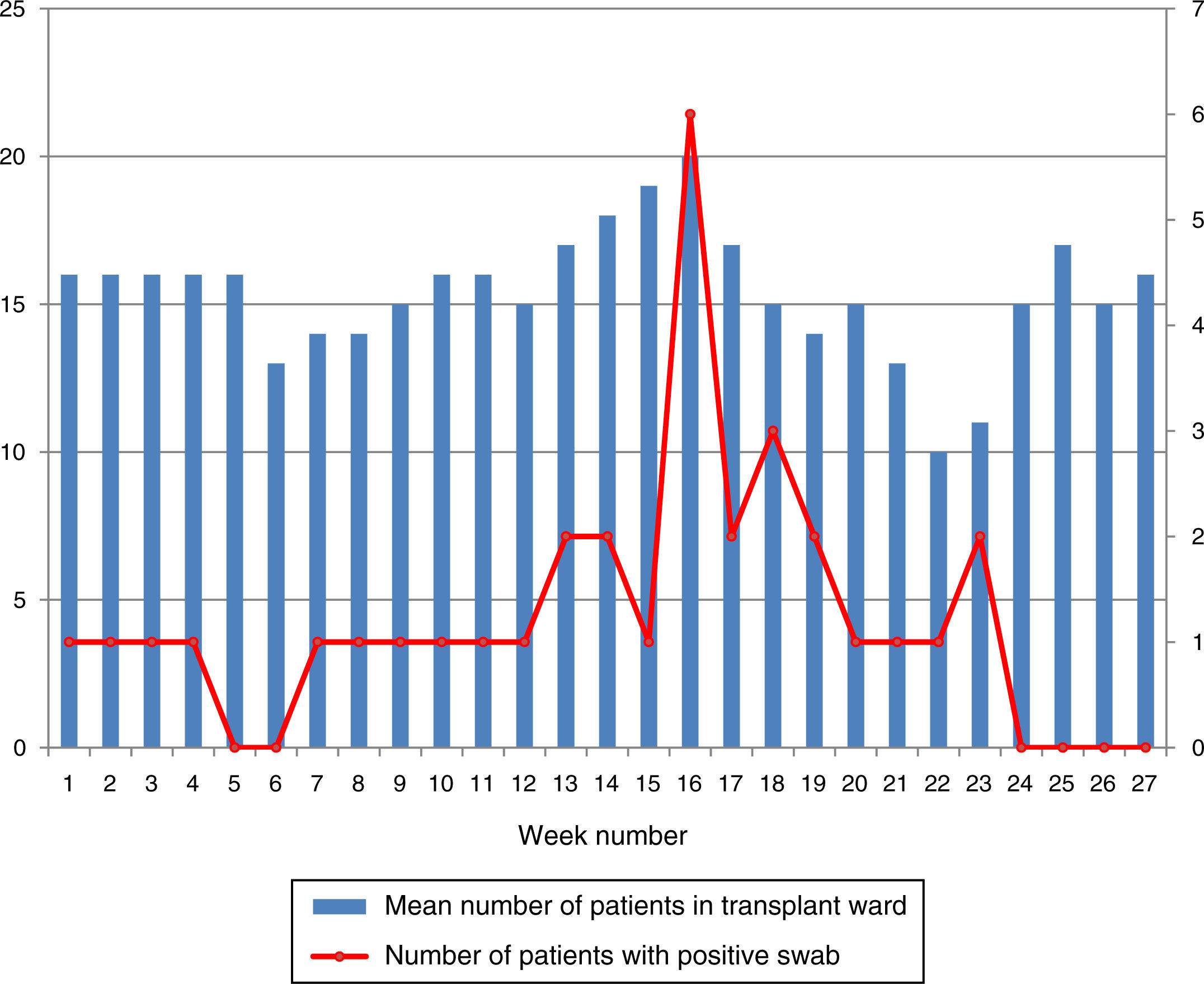
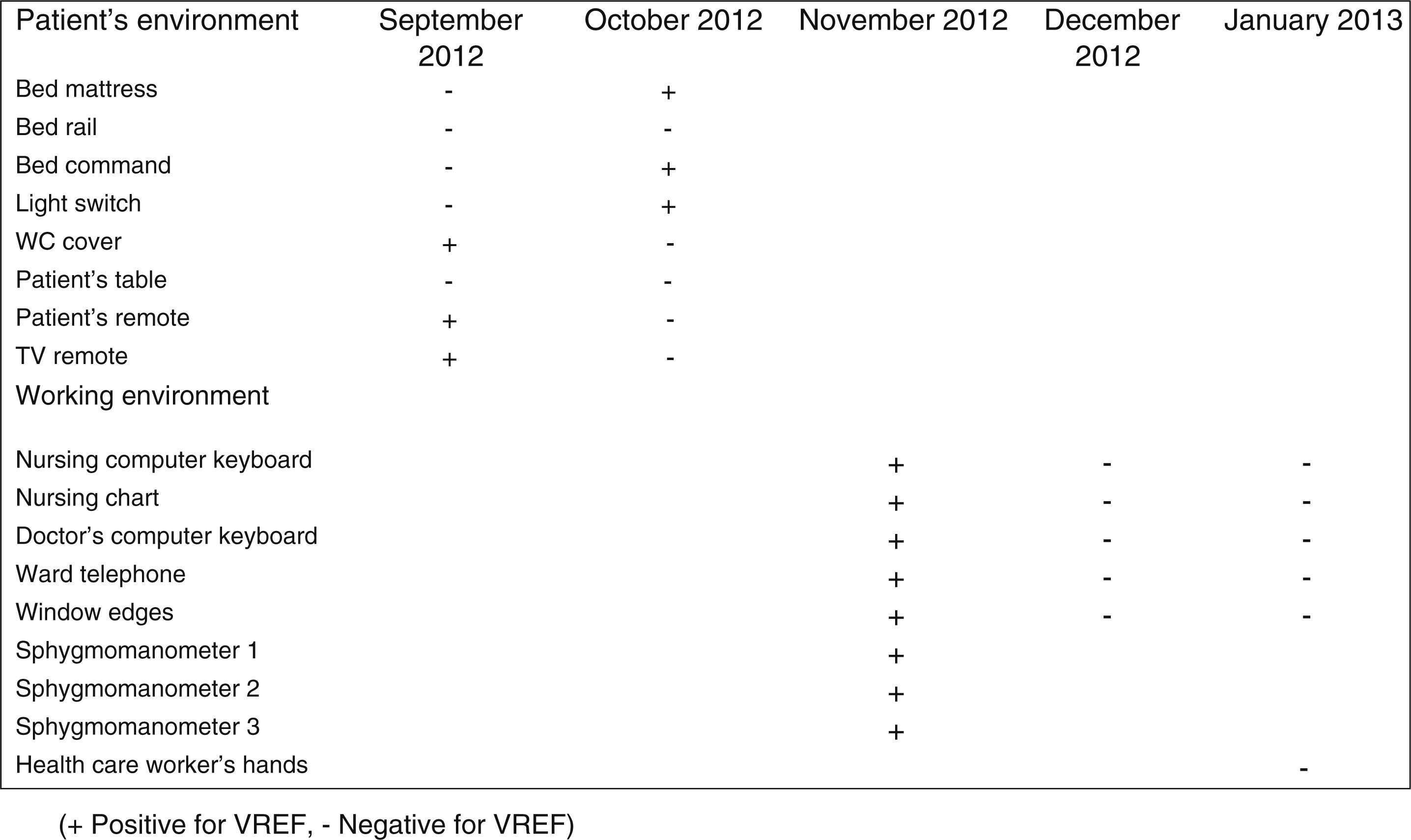
During the outbreak, 628 rectal swabs overall were collected from 151 patients (1–8 swabs/patient). VREF was detected in 74 swabs from 13 patients. Fig. 3 shows the weekly colonization pressure in the unit. There were no cases of infection among the studied patients and all were therefore considered as colonized. At the beginning of the outbreak, environmental (Fig. 4) cultures were performed on furniture, TV controls, WC cover and light switches in the patient's room. Afterwards, since the number of colonized patients continued to increase, environmental surveillance swabs were extended and standardized to other medical equipment and surfaces in the ward where many positive cultures were found on many devices, such as sphygmomanometers, stethoscopes, keyboards, computers and on working surfaces outside patient's rooms. After implementation of CDC Tier 2 measures, the environmental surveillance swabs were negative and no further positive cultures were found from December 2012 onwards. Fig. 2 illustrates the results of the second tier measures in patients: 7 cases were found in November, when first tier measures were applied and these dropped to 2 cases in December, one case in January and none after that, when second tier measures were implemented. All patients were tracked while the source of the outbreak was sought without success. The first case had several risk factors: lived in a long-term facility, was in hemodialysis program 3 times/week, had been admitted to the hospital 6 times in the last year, and had received several antibiotic treatments (including gentamycin) due to chronic ulcers in the lower limbs; it was suspected that this patient might have been the index patient.
Time series were not calculated since no interventions regarding antibiotic policy were made, nevertheless an analysis was made regarding antibiotic consumption at the renal transplant ward from the previous year and onwards. We found there was a significant increase in the use of aminoglycosides (Spearman's Rho coefficient: 0.647, p<0.001) and a significant decrease in the use of penicillin (Spearman's Rho coefficient: 0.681, p<0.001) after the outbreak, but not before. There were no significant changes in the consumption of carbapenems or cephalosporines before, during and after the outbreak.
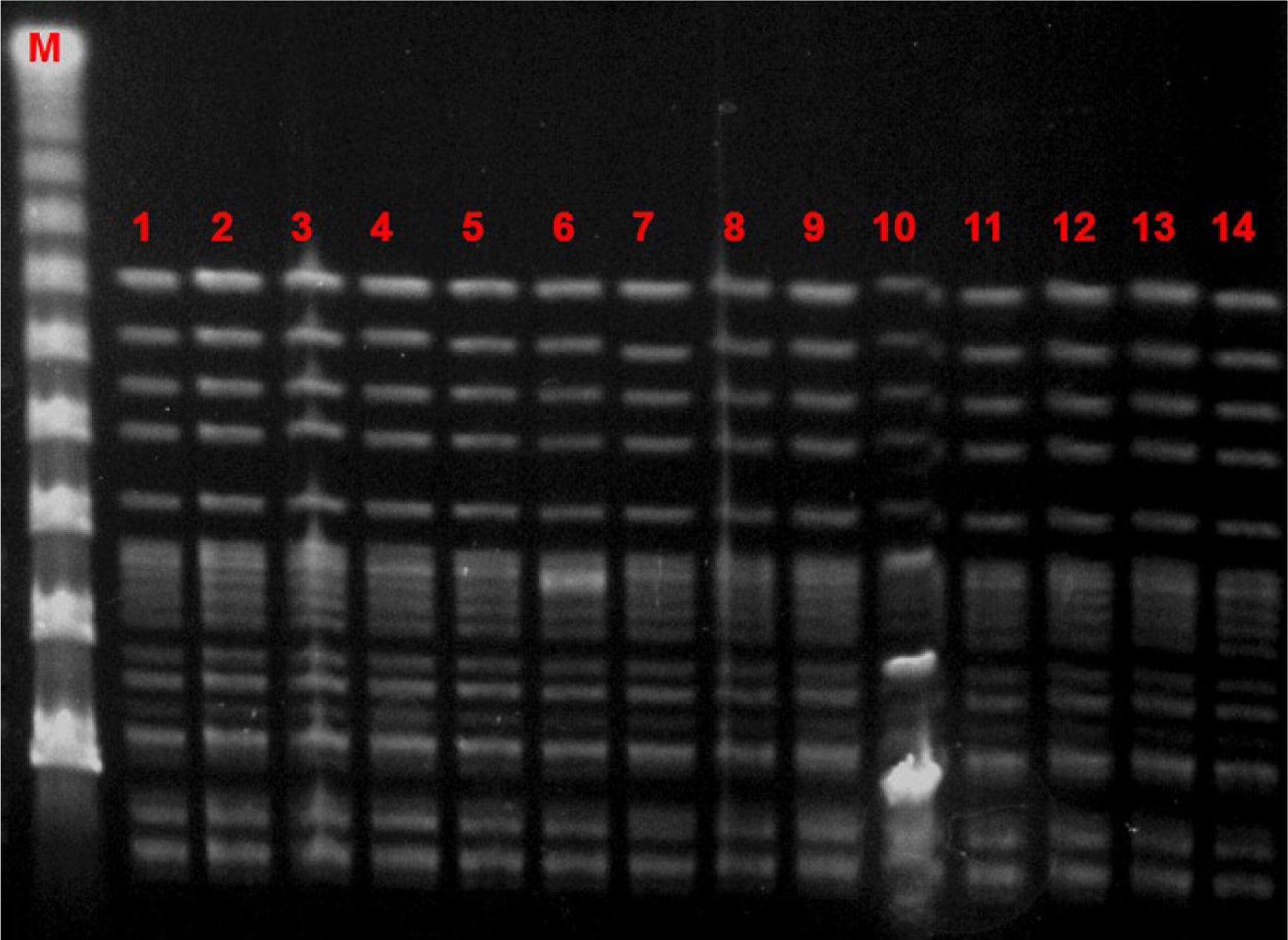
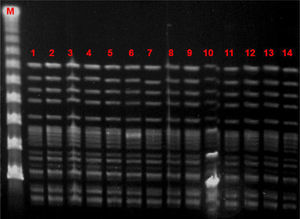
Microbiological characteristic of VREF isolatesThe VREF isolates clustered into a single pulsotype, designated PFGE-A, corresponding to ST17 (Fig. 5). Environmental isolates also belonged to the outbreak clone. All isolates carried the vanA gene and exhibited high-level resistance (MIC> 16mg/L) to vancomycin and teicoplanin. Moreover, all isolates were ampicillin-resistant (MIC >8mg/L) and high-level streptomycin- and gentamicin-resistant, but susceptible to linezolid and tetracyclines. Detection of virulence genes showed the presence of hyl genes in all isolates.
DiscussionVancomycin-resistant E. faecium and enterococci in general are tremendous colonizers, found in the majority of cases in the gastrointestinal tract.18 They have great genome plasticity and a tendency to environmental persistence, which enables transmission as well as the spread of resistance elements.19 The hands of health workers have proven to be the most significant means of transmission, since VREF can survive for up to 60min on the hands and up to 4 months on inanimate surfaces.20 In our series, first tier infection control measures were implemented, mainly targeting person-to-person transmission. When we realized that these measures were insufficient because the number of colonized patients continued to rise, environmental cultures were increased, which demonstrated extensive growth of VREF, not only in the patients’ rooms and on furniture but also on ward surfaces and medical equipment. No specific policies regarding the consumption of cephalosporins or glycopeptides were applied during this outbreak. Other series have demonstrated that environmental surfaces are a large reservoir for VREF in an outbreak scenario. In the outbreak published by Livornese et al.,21 VREF was isolated from the rectal probe handles of three electronic thermometers used only on non-isolated patients in their unit. Molecular epidemiology showed that all clinical and environmental isolates belonged to the same high-risk clone (ST17).
It was only after the environmental source was identified that the outbreak was controlled. We demonstrated that there was a massive presence of VREF in the ward. It was when second-tier infection control measures were applied (including universal private rooming and ceasing new admissions to the unit) and the cleaning of surfaces and equipment was strongly reinforced that we were able to control the outbreak. Environmental surveillance cultures were proof of this since cultures were consistently negative after second-tier measures were implemented. As a result, the number of colonized patients dropped sharply. We believe that the implementation of these measures was key to avoiding further colonization and infection. Previous outbreaks in similar scenarios have reported a colonization/infection rate of 11%.22 The fact that no infections were reported in our series might be a result of the prompt outbreak control. Even if colonization is the most important factor for developing an infection, it is not the only one. Although the source of the outbreak was not found, we believe that poor hand hygiene and insufficient surface cleaning were the origin of the outbreak and key to the spread of VREF.
Several outbreaks have been reported in different countries across Europe in recent years, most of which occurred in the ICU or involved immunosuppressed patients.23–27 In Spain, epidemic outbreaks of VREF are rare, although some have been reported.28,29 The main genotypic pattern was carriage of the vanA gene,30,31 although vanB has also been reported. In our outbreak, the only gene detected was vanA. PFGE analysis revealed the dissemination of a single clone assigned by MLST to ST17, belonging to the previously designated clonal complex 17 (CC17). CC-17 has been defined as a hospital-adapted E. faecium subpopulation exhibiting ampicillin- and quinolone-resistance and, occasionally, vancomycin resistance.32 Furthermore, these hospital-derived strains are enriched in putative virulence genes such as esp and hyl, which may play a role in the adaptation of this genetic lineage to the hospital environment.32 Variable contents of esp and hyl genes have been described among vancomycin-resistant E. faecium strains, usually with the more prevalent esp gene.29 In our study, on the other hand, the outbreak clone carried only the hyl gene.
Several factors should be taken in account for the management of future outbreaks; firstly, previous studies have shown low sensitivity with single rectal swabs,33 which is the reason why rectal swabs were, and should be, repeated weekly during hospitalization. Secondly, the risk factors for prolonged carriage of VREF, such as surgery, antibiotic use during admission, dialysis, and discharge to a nursing home or other health care institution, should be taken into account,34 especially for patients readmitted to the hospital in the months following discharge.
Interestingly, in our outbreak there were no cases of infection, so that we were not faced with the challenge of treating a very complicated group of patients with limited therapeutic options. The limitations of our study are that it is a case series without controls, and even if interventions were done, it was not in a controlled study scenario. Other limitation regards the screening for VREF, using a rectal swab that was seeded in Campylobacter blood agar plates; the low density of this microorganism in the feces justifies a pre-enrichment in BHI broth supplemented with 4–6mg/L of vancomycin and further growth in selective media (i.e. m-Enterococcus agar plates) supplemented also with vancomycin. This is probably the best way to recover all positive carriers, and we might have missed some carriers.
In conclusion, a clonal outbreak of VREF-ST17 in Spain involving colonization only is reported. The prompt implementation of aggressive infection control measures, highlighting strong environmental surveillance and more directed and effective cleaning, was efficacious in controlling the outbreak and probably avoided the emergence of infect patients.
Ethical approvalAll procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent: For this type of study formal consent is not required.
FundingNo funding was provided for this study.
Conflict of interestThe authors declare no conflicts of interest.