Mutations in the SARS-CoV-2 genome can affect the gene encoding the Spike (S) antigen, which interacts with the host cell specific receptor, selecting mutant variants with changes in their infective capacity, pathogenic potential and resistance to neutralizing antibodies. The nomenclature to design the variants uses a colloquial form referred to the country or place of detection, a code from the “Pangolin” database and one from the “Nextstrain” page. New variants that have spread include the British B.1.1.7 (20I/501Y.V1), the South African B.1.351 (20H/501.V2), the Brazilian P.1 (20J/501Y.V3), the Californians B.1.427 B.1.429 (20C/S:452R) and the most recent, the Indian B.1.617 (VUI-21APR-01).
The gold standard for the identification of the variants is whole genome sequencing. However, real-time PCR techniques have already been developed for the detection of specific mutations that can facilitate their presumptive identification.
The impact of these variants on global vaccination programs has raised concern. It is generally thought that, since the response evoked by the vaccine against the S antigen is directed at the entire protein and the mutations only affect specific regions, the escape effect of the vaccine antibodies will be limited. Among the future strategies proposed for immuno-protection, the increase in the number of doses, the alternation of vaccines and the development of specific vaccines against different variants has been suggested.
Las mutaciones en el genoma de SARS-CoV-2 pueden afectar al gen que codifica el antígeno espicular (S), que interactúa con el receptor específico de la célula huésped, seleccionando variantes mutantes con alteraciones en su capacidad infectiva, potencial patógeno y resistencia a los anticuerpos neutralizantes. En la nomenclatura de las variantes se utiliza una forma “coloquial” que suele hacer referencia al país o lugar de detección, un código de la base de datos “Pangolín” y uno de la página “Nextstrain”. Entre las nuevas variantes que se han ido propagando se incluyen la Británica B.1.1.7 (20I/501Y.V1), la Sudafricana B.1.351 (20H/501.V2), la Brasileña P.1 (20J/501Y.V3), las Californianas B.1.427 y B.1.429 (20C/S:452R) y la más reciente, la India B.1.617 (VUI-21APR-01).
El método de referencia para la identificación de las variantes es la secuenciación del genoma completo. Sin embargo, ya se han desarrollado técnicas de PCR en tiempo real para la detección de mutaciones específicas que pueden facilitar su identificación presuntiva.
El impacto de estas variantes en los programas globales de vacunación ha suscitado inquietud. En general se piensa que, dado que la respuesta evocada por la vacuna frente al antígeno S se dirige a la totalidad de la proteína y las mutaciones sólo afectan a regiones concretas, el efecto de escape a los anticuerpos vacunales será sólo limitado. Entre las estrategias futuras propuestas para la inmuno-protección se ha sugerido el incremento del número de dosis, la alternancia vacunal y el desarrollo de vacunas específicas frente a diferentes variantes.
Although the SARS-CoV-2 genome appears relatively stable, this type of single-stranded RNA virus accumulates an estimated mutation rate of around 10−6–10−4 per replicative cycle. These mutations can affect the interaction of the spike antigen (S) with the host cellular receptor (angiotensin-converting enzyme 2 [ACE2]), affecting susceptibility to immune response, the aggressiveness of the infection, and/or the transmission capacity of the virus. Thus, some mutations may confer evolutionary advantages that favour the selection of certain lineages or variants.1
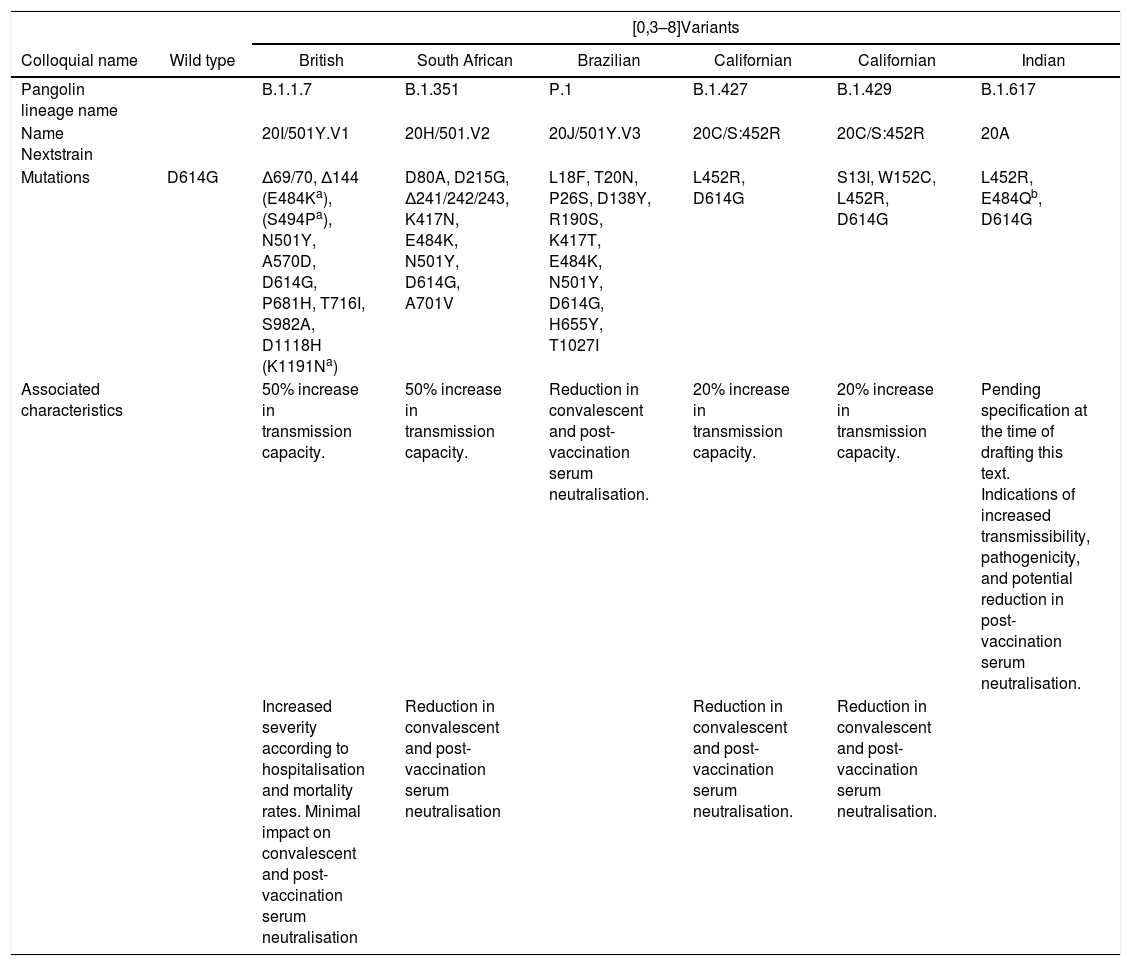
Variants can appear from genetic deletions (characterised by the loss of a portion of the nucleic acid sequence), synonymous mutations (also known as silent or nonsense mutations, which do not cause changes in amino acid synthesis) or non-synonymous mutations (associated with a change in amino acid coding). The latter are the most relevant functionally. Mutations that are termed “escape” subsequently expand under selective immunological pressure that limits, but does not eliminate, replication.2 Different variants have emerged in recent months that are co-circulating worldwide, and new variants are predicted to continue to emerge in the future. Table 1 summarises the main SARS-COV-2 variants currently circulating.
Main SARS-COV-2 variants currently circulating.7
| [0,3–8]Variants | |||||||
|---|---|---|---|---|---|---|---|
| Colloquial name | Wild type | British | South African | Brazilian | Californian | Californian | Indian |
| Pangolin lineage name | B.1.1.7 | B.1.351 | P.1 | B.1.427 | B.1.429 | B.1.617 | |
| Name Nextstrain | 20I/501Y.V1 | 20H/501.V2 | 20J/501Y.V3 | 20C/S:452R | 20C/S:452R | 20A | |
| Mutations | D614G | Δ69/70, Δ144 (E484Ka), (S494Pa), N501Y, A570D, D614G, P681H, T716I, S982A, D1118H (K1191Na) | D80A, D215G, Δ241/242/243, K417N, E484K, N501Y, D614G, A701V | L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I | L452R, D614G | S13I, W152C, L452R, D614G | L452R, E484Qb, D614G |
| Associated characteristics | 50% increase in transmission capacity. | 50% increase in transmission capacity. | Reduction in convalescent and post-vaccination serum neutralisation. | 20% increase in transmission capacity. | 20% increase in transmission capacity. | Pending specification at the time of drafting this text. Indications of increased transmissibility, pathogenicity, and potential reduction in post-vaccination serum neutralisation. | |
| Increased severity according to hospitalisation and mortality rates. Minimal impact on convalescent and post-vaccination serum neutralisation | Reduction in convalescent and post-vaccination serum neutralisation | Reduction in convalescent and post-vaccination serum neutralisation. | Reduction in convalescent and post-vaccination serum neutralisation. | ||||
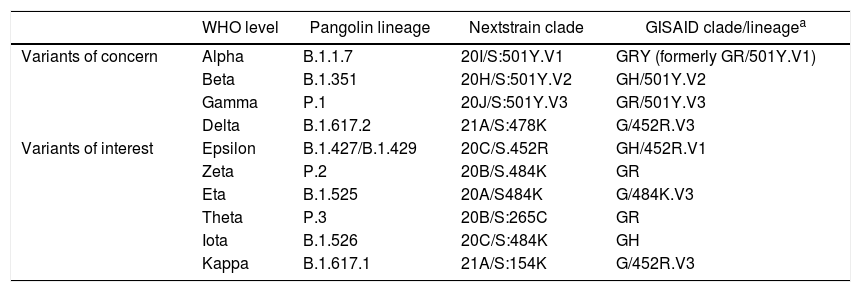
Although the terms strain and variant are usually used synonymously after mutations have occurred, in a pure sense they should cover the induction or otherwise of changes in the behaviour of the virus.3 However, these terms will be used interchangeably in this text. The nomenclature of the different variants of SARS-CoV-2 that have gradually emerged uses a “colloquial” form that usually refers to the country or place where the variant was detected (British strain), a code from the Pangolin database4 (B.1.1.7), and one based on the Nextstrain page5 (20I/501Y.V1). This multiple system can often lead to confusion. To provide information, the World Health Organisation (WHO) very recently adopted a simpler system based on letters of the Greek alphabet to denote the various variants.6Table 2 shows the new nomenclature for the different variants.
WHO nomenclature, proposed for naming variants of concern and variants of interest.
| WHO level | Pangolin lineage | Nextstrain clade | GISAID clade/lineagea | |
|---|---|---|---|---|
| Variants of concern | Alpha | B.1.1.7 | 20I/S:501Y.V1 | GRY (formerly GR/501Y.V1) |
| Beta | B.1.351 | 20H/S:501Y.V2 | GH/501Y.V2 | |
| Gamma | P.1 | 20J/S:501Y.V3 | GR/501Y.V3 | |
| Delta | B.1.617.2 | 21A/S:478K | G/452R.V3 | |
| Variants of interest | Epsilon | B.1.427/B.1.429 | 20C/S.452R | GH/452R.V1 |
| Zeta | P.2 | 20B/S.484K | GR | |
| Eta | B.1.525 | 20A/S484K | G/484K.V3 | |
| Theta | P.3 | 20B/S:265C | GR | |
| Iota | B.1.526 | 20C/S:484K | GH | |
| Kappa | B.1.617.1 | 21A/S:154K | G/452R.V3 |
The different mutations associated with changes in amino acids are named with an alpha-numeric key that indicates, firstly, the code of a capital letter of the amino acid that has been replaced, a number that refers to the position of the change and finally the capital letter corresponding to the code of the new amino acid. Thus, for example, the mutation N501Y indicates that the amino acid asparagine (N) has been replaced by a tyrosine (Y) at position 501.
Classification of the SARS-CoV-2 variantsThe different SARS-CoV-2 variants have been classified according to their level of importance7:
Variants of Interest (VOI) are those that have been associated with changes to receptor binding, reduced neutralisation by antibodies generated against previous natural infection or vaccination, reduced efficacy of treatments, potential diagnostic impact, or predicted increase in transmissibility and/or disease severity, with clusters of cases and outbreaks, but with limited prevalence in other countries. Examples of variants of interest are B.1.525 (20A/S:484K) B.1.526 (20C/S:484K), B.1.526.1 (20C), B.1.617 (20A), B.1.617.1 (20A/S:154K), B.1.617.2 (20A/S:478K), B.1.617.3 (20A), and P.2 (20J).7
Variants of Concern (VOC) include variants where there is evidence of an increase in transmissibility, severity, reduction in neutralisation by natural antibodies or those generated by vaccination, reduced effectiveness of treatments or diagnostic test failures. Examples of variants of concern are the British B.1.1.7 (20I/501Y.V1), the South African B.1.351 (20H/501.V2), the Brazilian P.1 (20J/501Y.V3), the Californian B.1.427 (20C/S:452R) and the Californian B.1.429 (20C/S:452R).7
Variants of High Consequence (VOHC) include those for which there is clear evidence that prevention and therapeutic measures have significantly reduced effectiveness relative to previously circulating variants. Fortunately, at present, there are no known SARS-CoV-2 variants that fall into this category.7
The European Centre for Disease Prevention and Control (ECDC) envisages an additional category, termed variants under monitoring, for those with similar properties to variants of concern, but for which there is only preliminary scientific evidence.8
Wild type strainThe original strain that started the COVID-19 pandemic in 2020 carried the 484E mutation. This mutation is located in the epitope of the dominant region encoding the S protein.9 Between January and February 2020, a SARS-CoV-2 variant emerged with a D614G substitution in the gene encoding the S protein.10 The original strain had the 614D precursor.11 This D614G mutation in the wild type strain enhanced its ability to drive the pandemic.12 Over a period of several months the D614G mutant strain replaced the strain initially identified in China and by June 2020 it had become dominant. This strain, compared to its precursor, showed increased infectivity and transmissibility. However, it was not associated with increased severity, impaired effectiveness of laboratory diagnostic tests, decreased neutralising capacity of antibodies or reduced therapeutic effectiveness.10
British variantThe variant termed the British B.1.1.7 (20I/501Y.V1) was detected at the end of 2020 in the UK and has rapidly spread to many countries,13 often supplanting the original wild type strain. It has more than 20 mutations, excluding those not associated with changes in amino acids, almost half of which affect the S antigen coding regions (the main target of neutralising antibodies).14 This variant maintains the D614G mutation and also shows the N501Y mutation that seems to increase the interaction of the S protein with the ACE2 receptor15 as it affects the receptor binding domain (RBD).16 In addition, it has the 69/70 deletion.10,16 This deletion may decrease the diagnostic sensitivity of certain PCR tests that target the gene coding for the S antigen. However, since most PCR techniques combine multiple targets, this may not be too problematic. These molecular changes seem not to impact the performance of rapid antigen tests.10 This variant has been associated with increased transmissibility and severity. Although the increased clinical severity of this and other variants may be questionable,10,17 the increased mortality reported for this strain is 4.1 deaths per 1000 cases detected (compared to that of previously circulating strains estimated at 2.5 per 1000 cases)18 and it shows relative resistance to neutralisation by monoclonal antibodies to the S antigen. However, it is not more resistant to plasma from individuals exposed to natural infection or following vaccination.19
South African variantThe B.1.351 variant (20H/501.V2),2,13,15 which may have appeared a few months earlier, was reported in South Africa in December 2020. This variant, which emerged independently of B.1.1.7.,16 harbours several non-synonymous mutations and a deletion that mostly affects the S protein.14 It combines the D614G and N501Y15 mutations with the K417N, E484K20 mutations. Unlike the British strain, it does not carry the 69/70 deletion.16 The N501Y mutation in the South African variant, although also present in the UK variant, appears to be phylogenetically distinct.10 There is uncertainty as to whether it is associated with greater severity or worse prognosis.10 Compared to the wild type strain, it appears to be markedly more resistant to neutralisation by serum from convalescent or vaccinated subjects.19,21,22
Brazilian variantThe Brazilian variant P.1 (B.1.1.28.1, 20J/501Y.V3)13 was detected in Japan from travellers from Brazil.16 It contains several mutations, including three affecting the S protein.14,16 The E484K mutation, shared by both the Brazilian and South African variants,23 has triggered alarm due to its possible link to some immune evasiveness.14 Both variants have been associated with increased transmissibility and a higher reinfection rate.13,14 The other two mutations of this variant that affect the S protein are K417T and N501Y.16 A way of distinguishing the South African variant from the Brazilian variant is that the Brazilian variant lacks the K417N mutation.
B.1.1.7 variant with E484K mutationThe detection of the E484K mutation in B1.1.7. sequences has also raised concerns about a loss of vaccine efficacy, due to its combination with the other mutations of the British variant.14
Californian variantOther emerging variants include strain B.1.429 (20C/S:452R)15,24,25 and B.1.427 (20C/S:452R)24 circulating in California. The B.1.429 variant, which has the L452R, D614G and W152C7,25 mutations, among others, has spread rapidly in the US and other countries, and is less susceptible to neutralisation by serum from convalescent or vaccinated subjects.21 These variants have been classified as variants of concern by the CDC (Centres for Disease Control and Prevention),7 whereas the WHO considers them only variants of interest.6
Indian variantMore recently, there has been widespread alarm about the emergence and spread in India of the new variant B.1.617,26 also known as VUI (Variant Under Investigation)-21APR-0127,28 and 20A according to “Nextstrain”.7 It is considered a variant of interest for the time being.7 Some media are reporting this strain as a “double mutant” because it combines the L452R mutations of the California strain with the E484Q mutation. The term “double mutant” is meaningless considering that the other SARS-CoV-2 variants also carry several mutations simultaneously. Other related lineages are B.1.617.1 (20A/S:154K), B.1.617.2 (20A/S:478K), and B.1.617.3 (20A). These three lineages have the L452R mutation in common. The variants B.1.617.1 and B.1.617.3 (both considered variants of interest by the CDC and the ECDC)7,8 share the E484Q7 mutation with B.1.617. The B.1.617.2 variant, although still classified as a variant of interest by the CDC,7 has recently been considered a variant of concern by the ECDC.8
It is thought that mutations in the S protein, which increase the transmissibility of the virus, could favour replacement of wild type precursor strains by the new variants already circulating and by potential future mutations.29
Variants and vaccinationThe impact of these variants on global vaccination programmes has raised concerns and the development of markers that correlate vaccine and protection is considered a priority.24 As we mention above, it has been suggested that the new variants could escape the immune response more easily. In general, it is considered that since the vaccine-evoked response to the S antigen targets the entire protein and mutations only affect specific regions, it does not appear that the new variants can completely evade the action generated after vaccination.3 The results of immunisation in Israel are encouraging in this regard, where the British variant is predominant.30 Preliminary trials have also shown an acceptable vaccine-induced neutralising response to different variants.28,31,32 The main SARS-CoV-2 vaccines include messenger RNA vaccines (mRNA-1273; Moderna and BNT162b2; Pfizer), vector vaccines (ChAdOx1; AstraZeneca and Ad26.COV2.S; Janssen), and the protein subunit vaccine NVX-CoV2373 (Novavax).33 Messenger RNA vaccines (mRNA-1273; Moderna and BNT162b2; Pfizer), vector vaccines (ChAdOx1; AstraZeneca and Ad26.COV2. Messenger RNA vaccines (mRNA-1273; Moderna and BNT162b2; Pfizer) appear to show only slightly lower efficacy against the UK variant. However, their neutralising capacity is significantly reduced against the South African variants and to a lesser extent against the Brazilian variant (given the high titre of antibodies generated for the latter variant, the decreased neutralising activity seems to have a lower impact than for the South African variant).33 It has recently been suggested that, although the Indian B.1.617.1 variant is also more resistant to neutralisation, the Moderna and Pfizer messenger RNA vaccines may well retain their ability to protect against it.34 The vector vaccine ChAdOx1 (AstraZeneca), although it may show a limited loss of activity against the British variant, maintains acceptable effectiveness against it. However, the efficacy of this vaccine against the South African variant seems to have been significantly reduced, and its use in certain countries with a high prevalence of infection by this variant has been questioned.33 This vector vaccine, despite producing an antibody response with a reduced neutralising capacity against the Indian B.1.617.1 variant, appears to maintain efficacy that limits the severity and mortality of the infection.35 The vector vaccine Ad26.COV2.S (Janssen) shows less than 60% efficacy for protection against moderate/severe disease caused by the South African strain.33 The protein subunit vaccine NVX-CoV2373 (Novavax) is more than 80% effective against the British variant but less than 50% effective against the South African variant.33 The Californian variants may behave in a very similar way to the British variant towards the vaccines.33
Existing data suggest that beyond the humoral immune response, the cellular response is capable of recognising the new variants of SARS-CoV-2.36 Thus, in addition to the production of specific antibodies, the vaccines could lead to the induction of a cellular response (helper T cells and cytotoxic T cells).33 The T-dependent immune response against the S protein of SARS-CoV-2 is characterised by high levels of interferon-gamma (IFN-ɣ) that can be detected by interferon-gamma release assays (IGRAs).37 These may be adjunct assays to determine immunostatus in the current pandemic.38
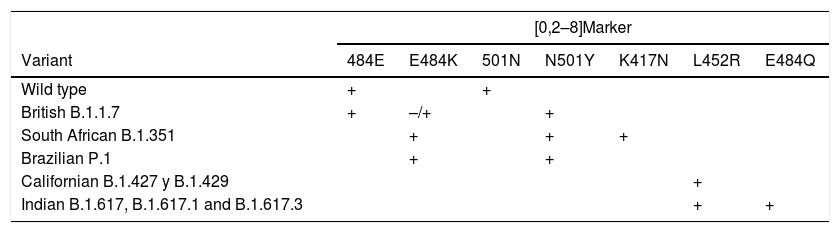
Diagnosis of variantsIn addition to the major clinical-epidemiological implications, a secondary consequence of the emergence of mutant strains is reduced performance of certain in vitro diagnostic tests.39 Whole genome sequencing methods are the gold standard for identifying SARS-CoV-2 variants.40 However, alternative, simpler and faster procedures based on real-time PCR techniques to detect specific mutations have recently been developed for this purpose.41 Certain combinations of mutant markers enable the main variants to be differentiated. The combination 484E and 501N identifies the wild type strain. The combination 484E and 501Y identifies the British strain. The 484K and 501Y combination occurs concomitantly in the Brazilian and South African variants, but the latter also carries the 417N mutation. The Californian variants contain the L452R mutation. The new Indian variants B.1.617, B.1.617.1 and B.1.617.3 carry the latter mutation together with E484Q. Table 3 shows the mutation markers that distinguish between the most important variants.
Markers of the mutations that enable differentiating between the main variants currently circulating.
| [0,2–8]Marker | |||||||
|---|---|---|---|---|---|---|---|
| Variant | 484E | E484K | 501N | N501Y | K417N | L452R | E484Q |
| Wild type | + | + | |||||
| British B.1.1.7 | + | –/+ | + | ||||
| South African B.1.351 | + | + | + | ||||
| Brazilian P.1 | + | + | |||||
| Californian B.1.427 y B.1.429 | + | ||||||
| Indian B.1.617, B.1.617.1 and B.1.617.3 | + | + | |||||
The mass vaccination that has begun in recent months may mark the beginning of the end of the current COVID-19 pandemic. However, it is foreseeable that new variants of the virus will continue to appear in the near future. This means that mutations must be monitored to adapt the relevant Public Health measures.10 Potential new emerging strains may entail changes in terms of their infectious capacity, clinical evolution, prognosis, and vaccine effectiveness. In the worst-case scenario, we should be vigilant for the potential and feared emergence of highly significant variants (as yet not identified). For the time being, standard preventive measures to reduce transmission, based on limiting interpersonal contact and maintaining safe social distances, remain in place.42 Certain vaccines retain their protective efficacy against new variants and thus their extensive use can be highly successful.40
To avoid the possibility of selection of vaccine escape variants, it is recommended that preparations be used that generate elevated levels of antibodies with high neutralising activity after only one dose and, for vaccines that require two doses, not to prolong for too long the interval between the first and second dose.33 Increasing the number of doses, alternating vaccines, and developing monovalent and polyvalent vaccines against different variants have been suggested as future strategies proposed for immunoprotection.43
FundingThe authors declare that they have received no funding for this study.
Conflict of interestsThe authors have no conflict of interests to declare.
Please cite this article as: Pérez-Abeledo M, Sanz Moreno JC. Variantes de SARS-CoV-2, una historia todavía inacabada. Vacunas. 2021;22:167–173.