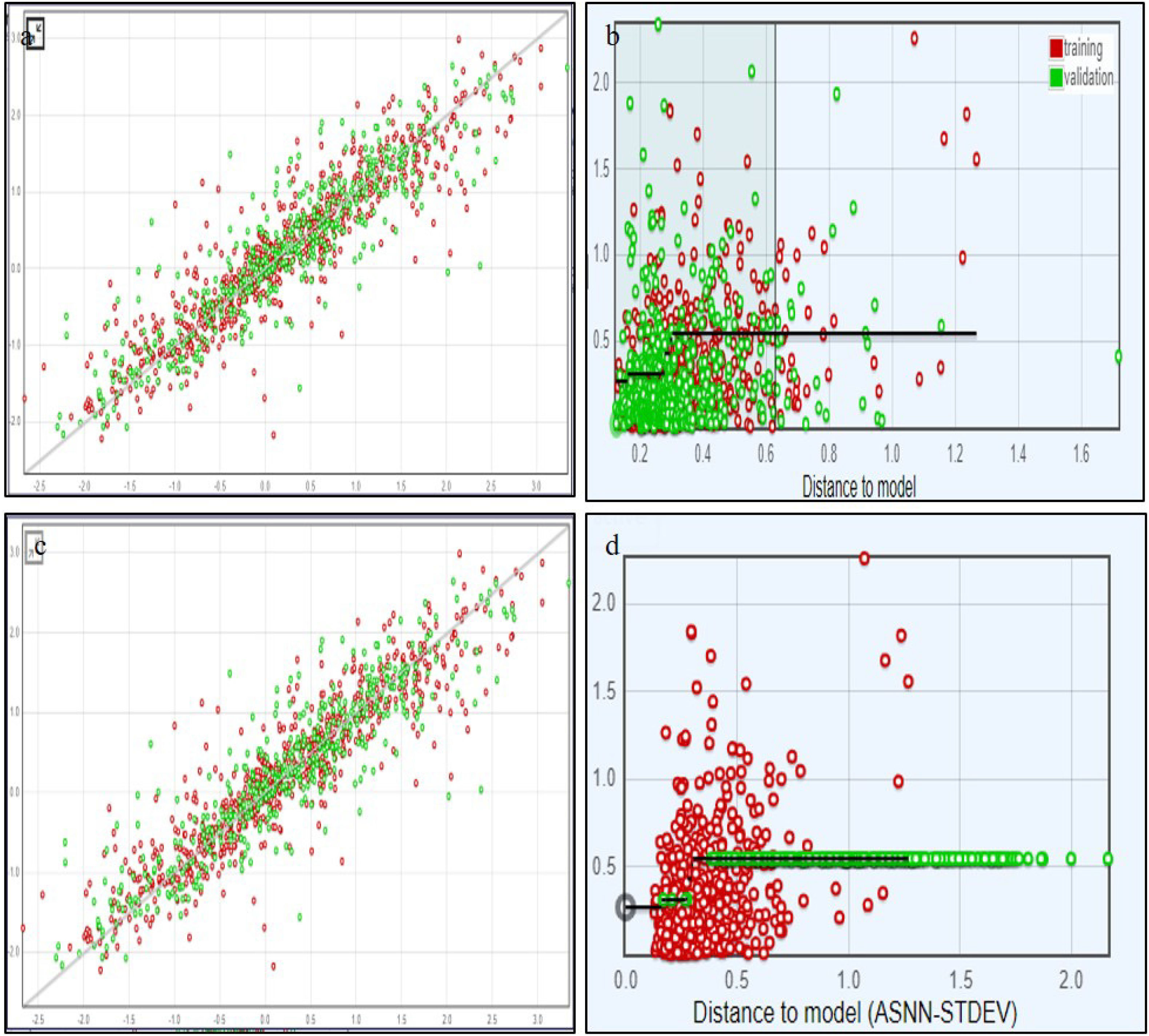
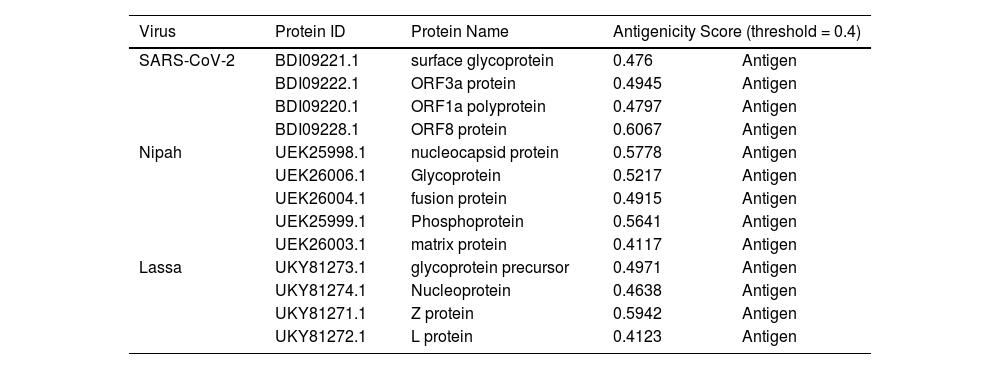
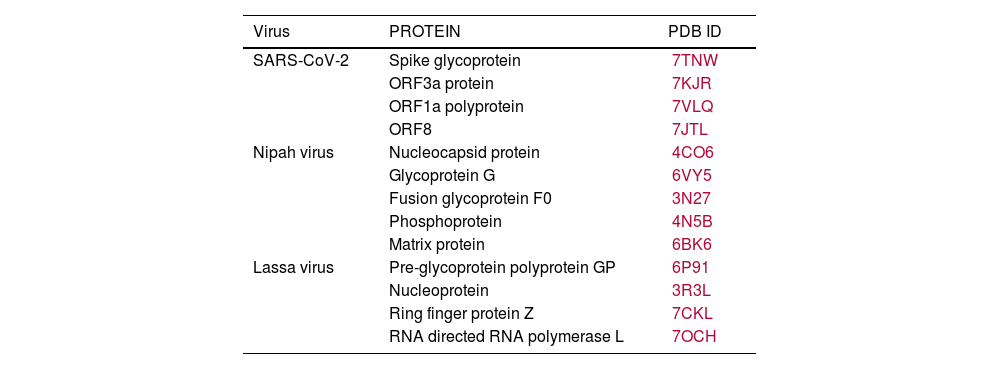
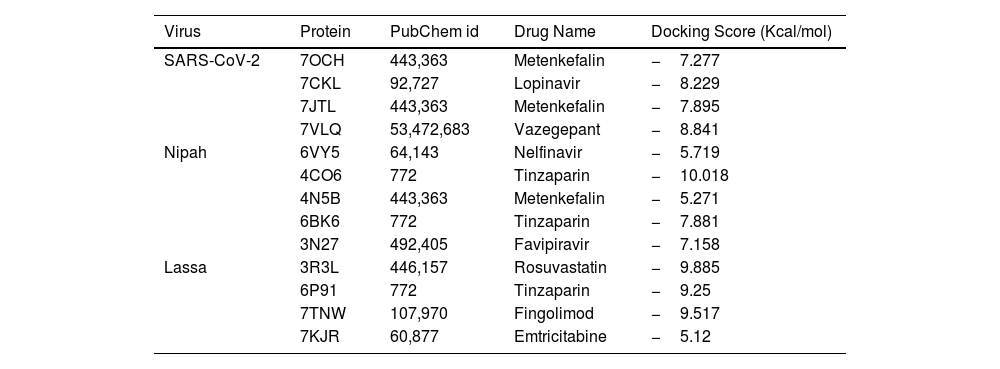
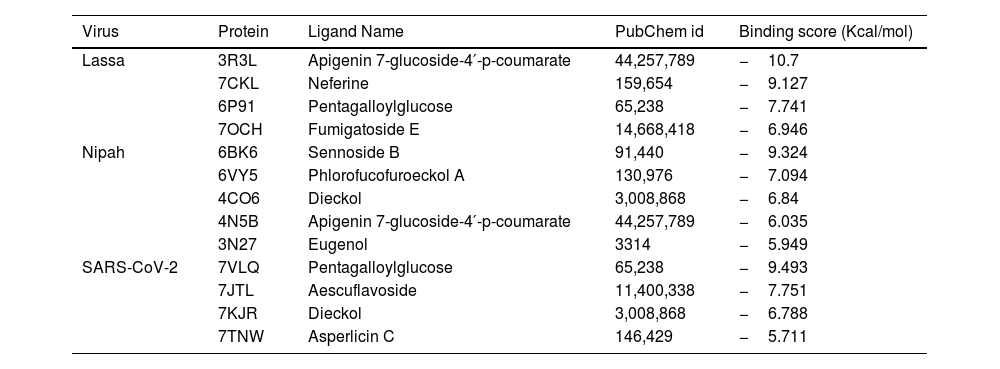
In recent years, public awareness of the risks posed by zoonotic diseases has significantly increased. Nipah virus (NiV) and Coronavirus are known to cause serious respiratory and neurological effects, while Lassa virus is a hemorrhagic virus. The current study employs an immunoinformatics approach to predict antigenic epitopes against NiV, Lassa virus (LASV), and Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) for the development of multi-epitope vaccines (MEV) and to identify the potential drug molecules using molecular docking approaches.
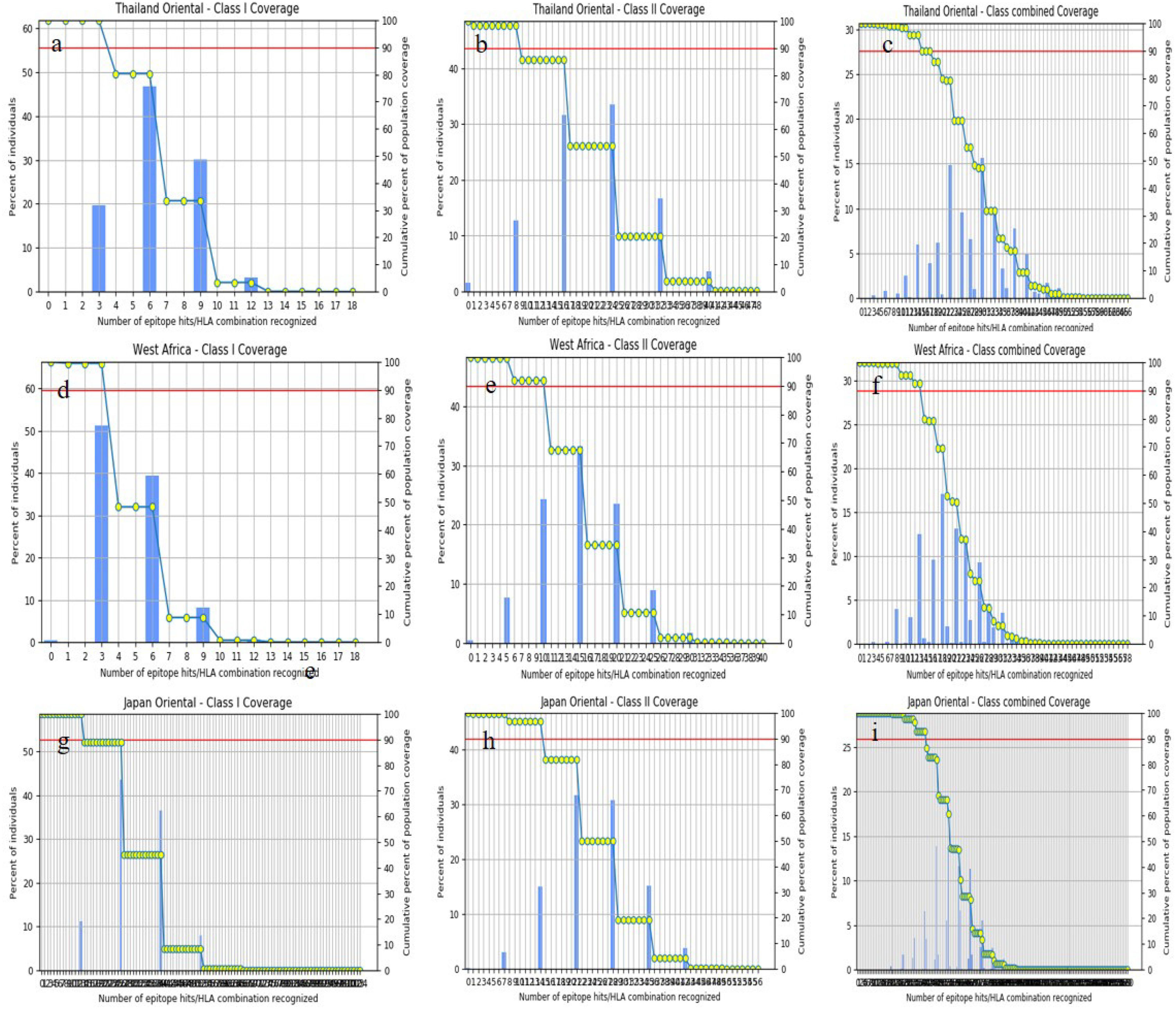
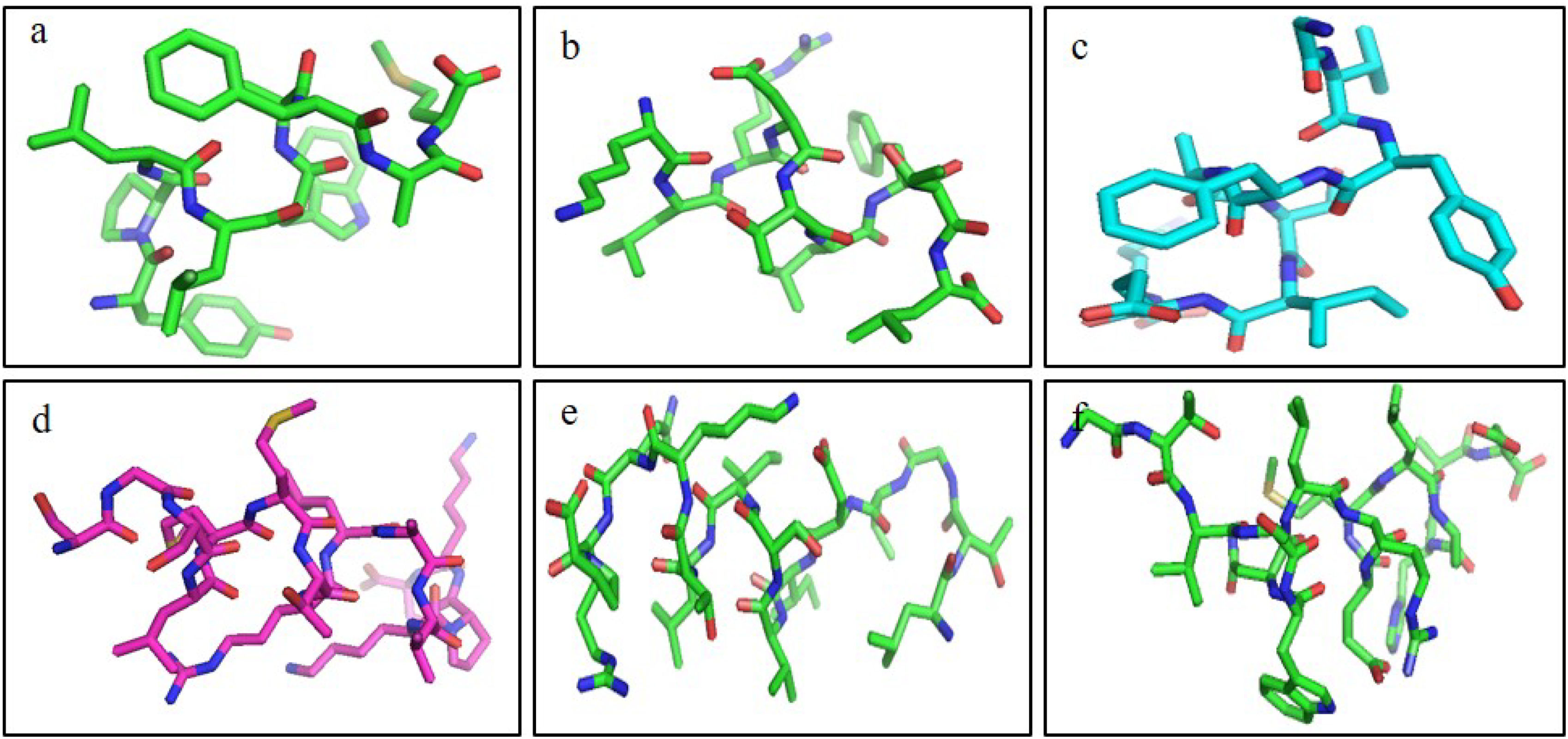
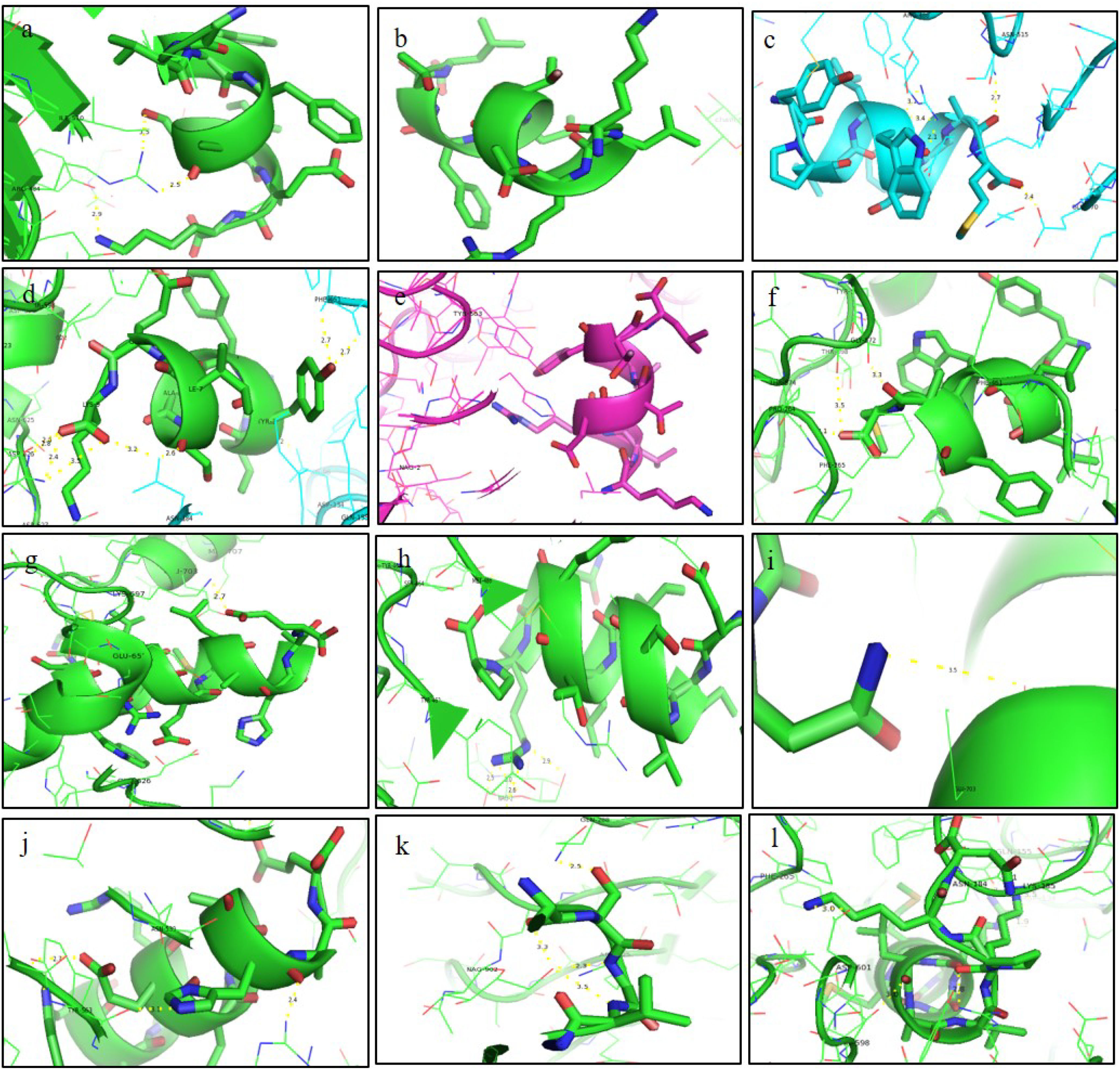
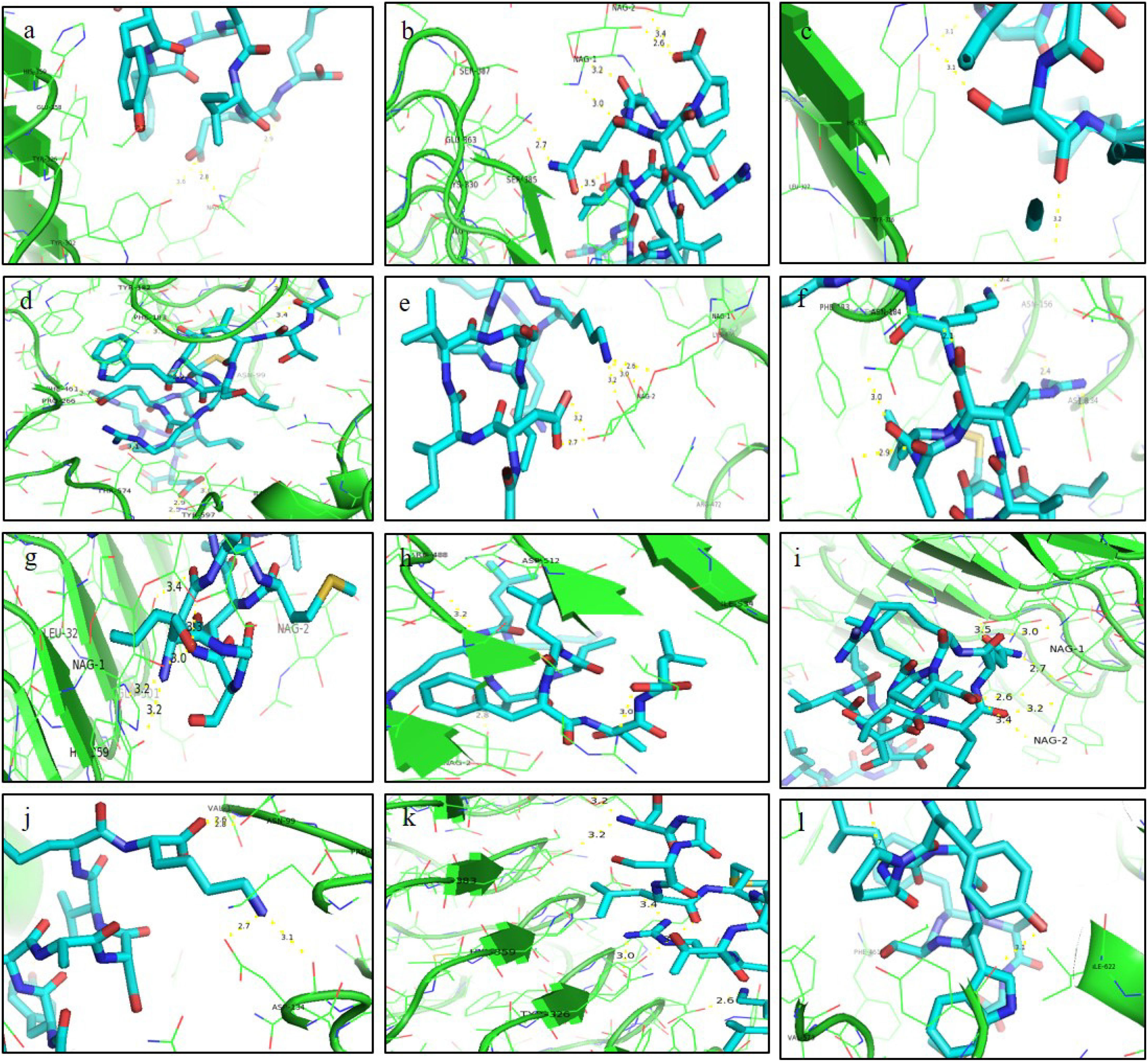
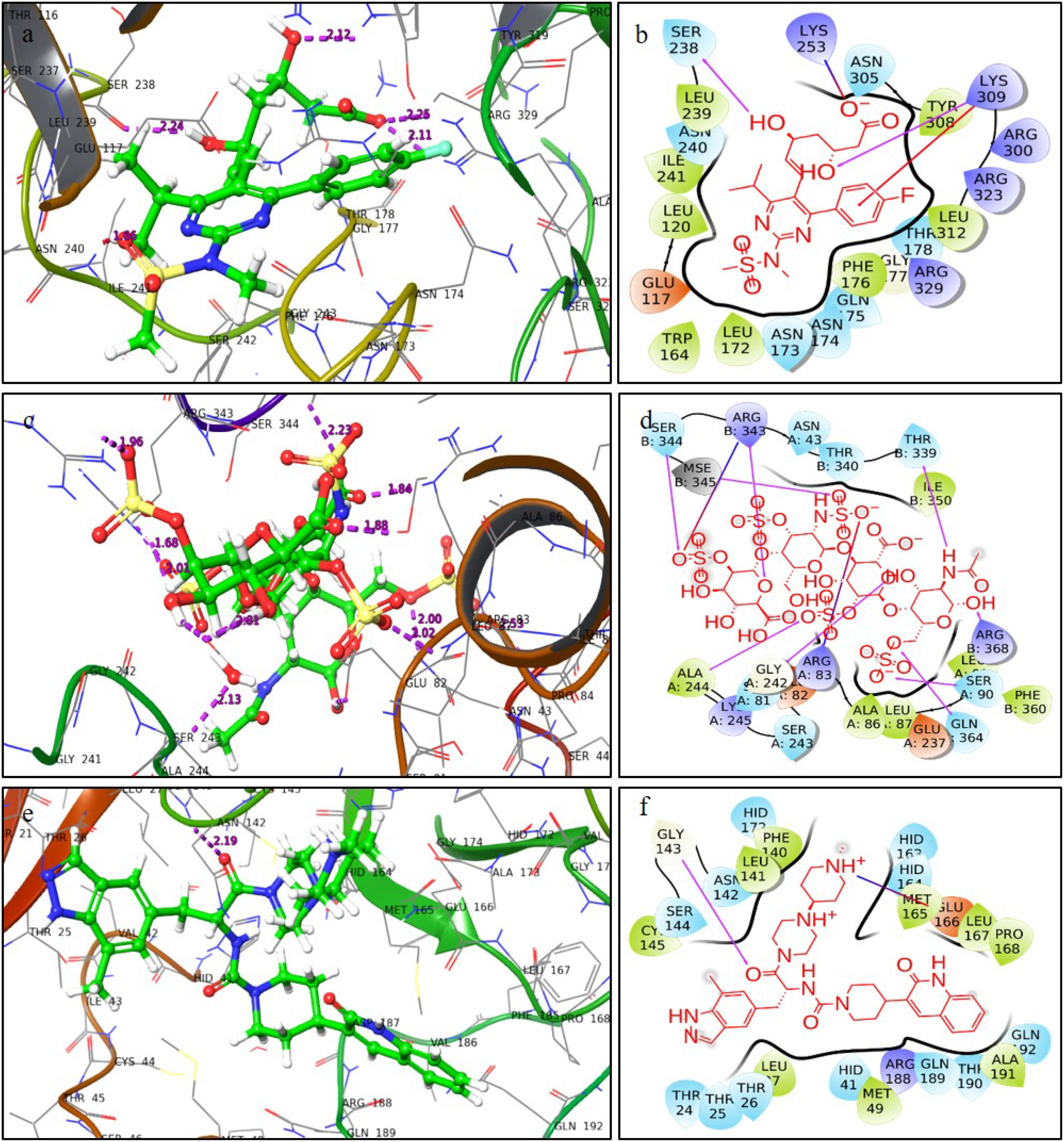
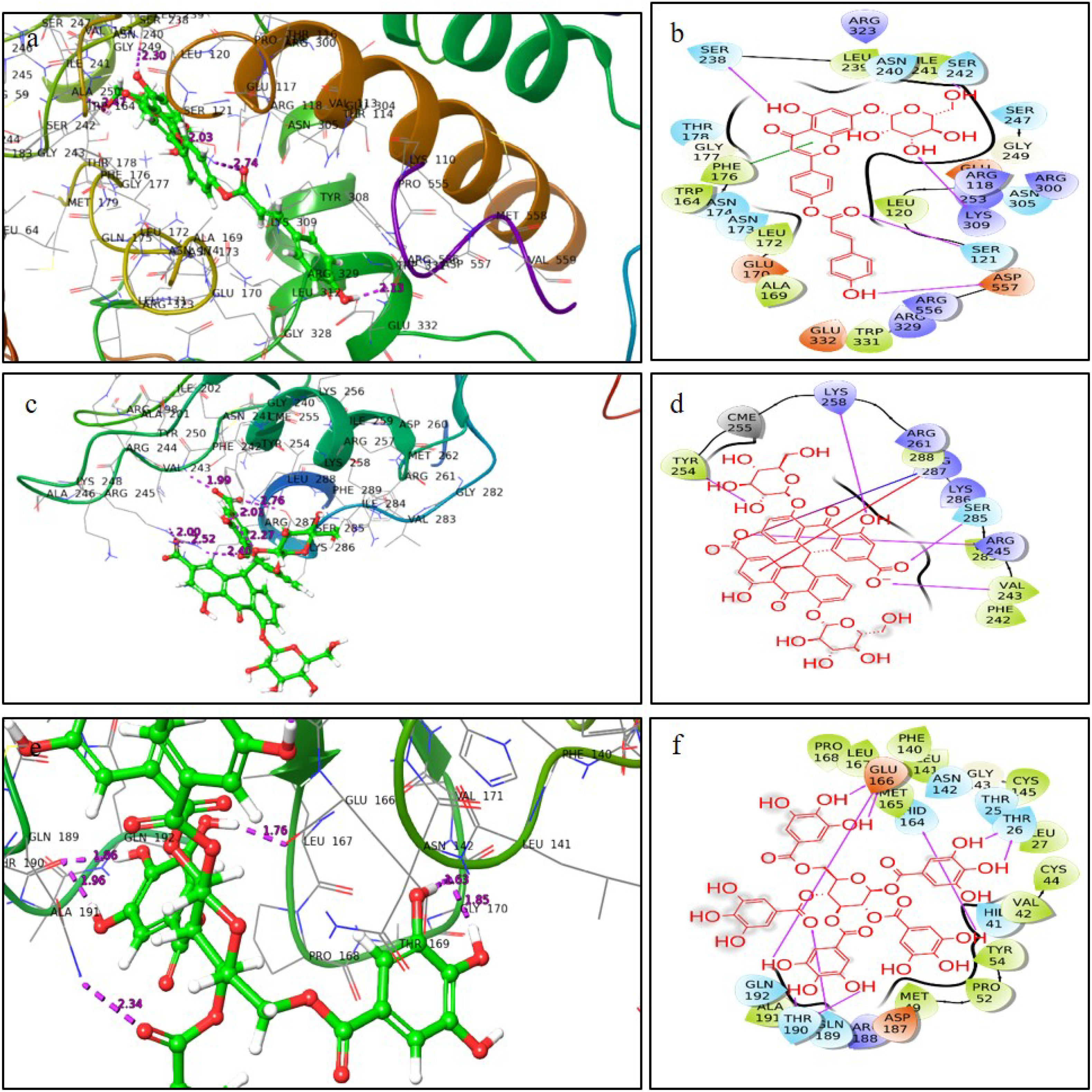
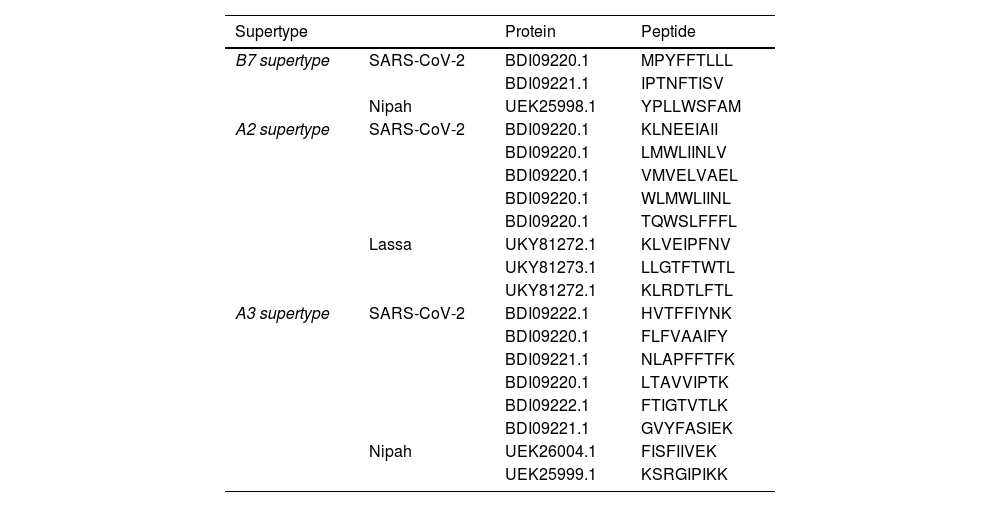
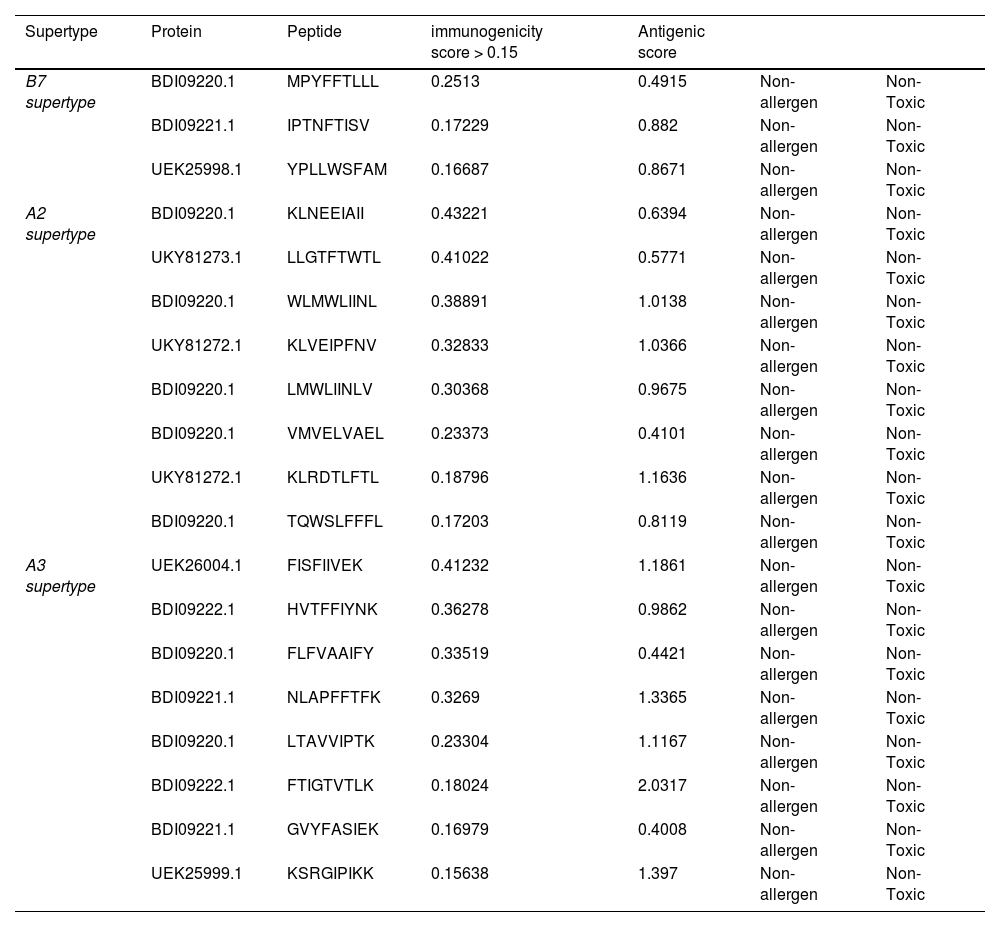
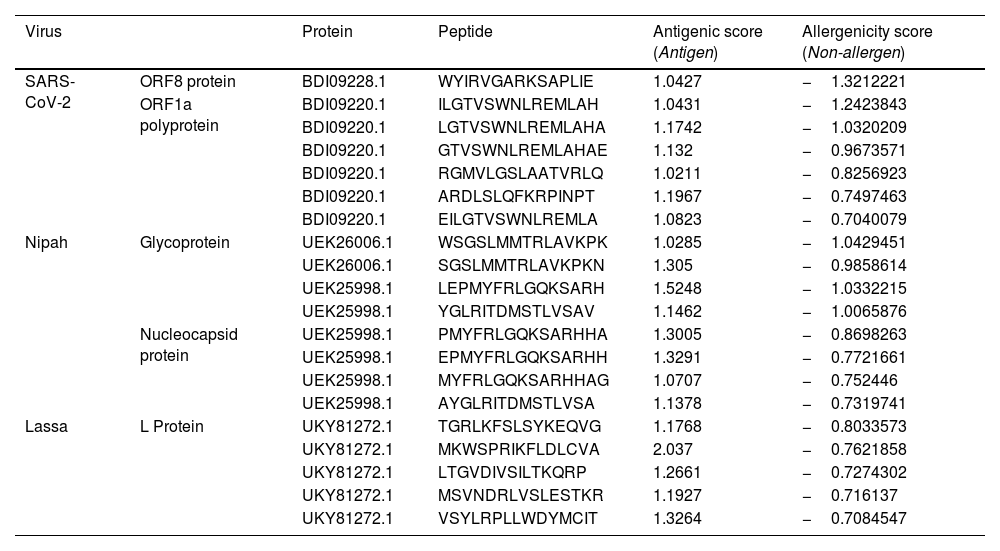
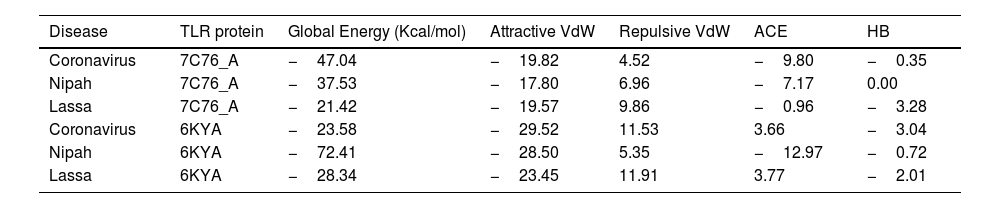
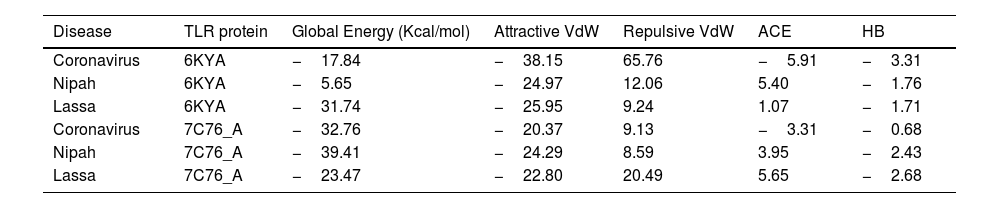
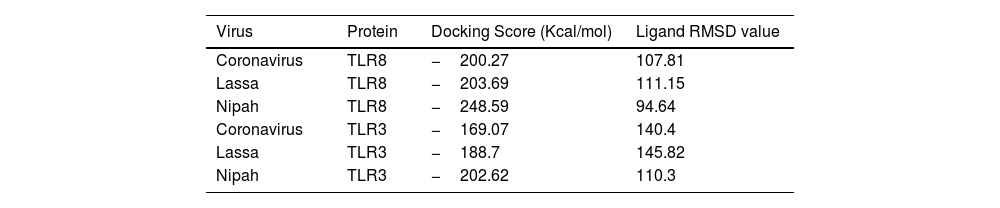
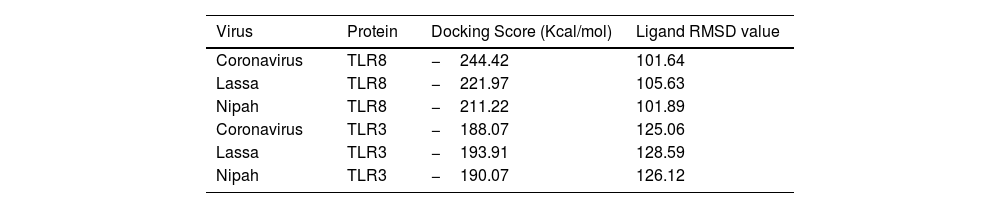
ResultsThirteen Cytotoxic T lymphocyte (CTL) epitopes for SARS-CoV-2, three for NiV, and three for LASV were selected. Additionally, eight Helper T lymphocyte (HTL) epitopes for NiV, seven for SARS-CoV-2, and five for LASV, all of which demonstrated antigenicity, non-allergenicity, and non-toxicity were identified and included. Molecular docking and subsequent construction of the 3D structure for the best epitopes from each virus revealed stable and strong binding affinities between the MEV and human pathogenic Toll-like receptors (TLRs), specifically TLR3 and TLR8.
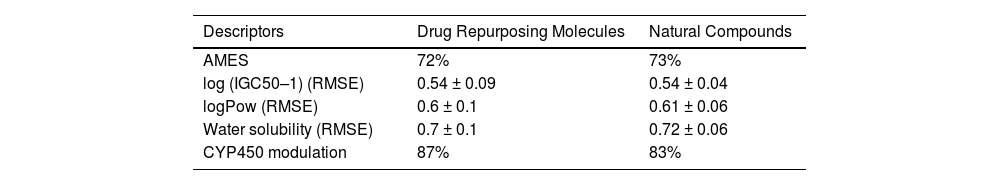
ConclusionThis work presents evidence of in silico research on vaccine design and molecular docking against the Nipah, Lassa viruses and SARS-CoV-2 target proteins. It highlights the computational approaches used for drug repurposing and the exploration of natural drug compounds. These findings suggest that the plant-derived or naturally sourced drugs exhibit significant potential in combating viral diseases in humans.
En los últimos años, ha incrementado considerablemente la concienciación pública sobre los riesgos planteados por las enfermedades zoonóticas. Se sabe que el virus Nipah (NiV) y el Coronavirus suscitan efectos respiratorios y neurológicos graves, mientras que el virus Lassa (LASV) es un virus hemorrágico. El estudio actual utiliza un enfoque inmunoinformático para predecir los epítopos antigénicos contra el NiV, el virus Lassa y el Coronavirus del Síndrome respiratorio agudo severo 2 (SARS-CoV-2) para el desarrollo de vacunas multiepitópicas (MEV), así como identificar las moléculas potenciales de los fármacos, utilizando técnicas de acoplamiento molecular.
ResultadosSe seleccionaron trece epítopos de linfocitos T citotóxicos (CTL) para SARS-CoV-2, tres para NiV, y tres para LASV. Además, se identificaron e incluyeron ocho epítopos de linfocitos T-helper (HTL) para NiV, siete para SARS-CoV-2, y cinco para LASV, demostrando todos ellos antigenicidad y ausencia de alergenicidad y toxicidad. El acoplamiento molecular y la construcción de la estructura en 3D subsiguiente de los mejores epítopos de cada virus revelaron afinidades de unión estables y fuertes entre las MEV y los receptores patogénicos humanos de tipo Toll (TLRs), específicamente TLR3 y TLR8.
ConclusiónEste estudio aporta evidencia de la investigación in silico sobre el diseño de vacunas y el acoplamiento molecular contra los virus Nipah y Lassa y las proteínas objetivo del SARS-CoV-2. Destaca los enfoques computacionales utilizados para la reutilización de fármacos y la exploración de compuestos farmacológicos naturales. Los hallazgos sugieren que los fármacos derivados de las plantas o de origen natural muestran un potencial significativo para combatir las enfermedades virales en humanos.