Adenoviruses play an important role in the etiology of severe acute lower respiratory infection, especially in young children. The aim of the present study was to evaluate the Human Adenovirus (HAdV) detection by different methods (Direct Fluorescence Assay – DFA and Nested Polymerase Chain Reaction – nested PCR), among samples collected from different groups of pediatric patients.
MethodsCollection of samples was made in children with congenital heart disease (CHD – 123 nasal aspirates collected in the years of 2005, 2007 and 2008) and in community children (CC – 165 nasal aspirates collected in 2008). Children were eligible if they presented acute respiratory infection (ARI) of probable viral etiology, within up to 7 days of symptoms' onset. All studied samples were evaluated by DFA and nested PCR assay.
ResultsOf the 290 samples included during the study period, 43 (14.8%) were positive on at least one test: 17/165 (10.3%) of the CC and 26/125 (20.8%) of the CHD children. The nested PCR detection rates in the community children were 15/165 (9.1%), and for children with CHD, 24/125 (19.2%). Molecular method showed higher detection rates when compared to the DFA test (p<0.001). Univariate analysis showed that children with congenital heart disease presented a significantly higher chance for acquiring the HAdV (Odds Ratio 2.3; 95% CI: 1.18-4.43).
ConclusionsBased on data obtained in the present evaluation, we suggest that a routine surveillance should be performed in high risk patients by molecular methods, thus improving diagnostic flow and efficiency.
Os adenovírus desempenham um papel importante na etiologia da infecção aguda grave do trato respiratório inferior, especialmente entre crianças. O objetivo do estudo foi avaliar a detecção do adenovírus humano (HAdV) por diferentes métodos (imunofluorescência direta – DFA e reação em cadeia da polimerase nested – nested PCR) em amostras coletadas de diferentes populações de pacientes pediátricos.
MétodosO material foi coletado de crianças portadoras de doença cardíaca congênita (DCC – 123 aspirados nasais coletados em 2005, 2007 e 2008) e de crianças da comunidade (CC – 165 aspirados nasais coletados em 2008). As crianças eram consideradas elegíveis se apresentas sem infecção respiratória aguda (IRA) de provável etiologia viral, com até sete dias de início dos sintomas. Todas as amostras coletadas no estudo foram avaliadas por meio de DFA e nested PCR.
ResultadosDe 209 amostras incluídas, 43 (14,8%) foram positivas em pelo menos um dos testes feitos: 17/165 (10,3%) das crianças da comunidade e 26/125 (20,8%) das crianças cardiopatas. As taxas de detecção por nested PCR foram 15/165 (9,1%) em crianças da comunidade e 24/125(19,2%) em crianças cardiopatas. O método molecular mostrou maiores taxas de detecção quando comparado com a DFA (p<0,001). A análise univariada mostrou que as crianças portadoras de cardiopatia congênita apresentaram chance significativamente maior de adquirir HAdV (odds ratio 2,3; IC 95%: 1,18-4,43).
ConclusõesBaseado nos resultados obtidos na presente avaliação, recomenda-se a vigilância de rotina em pacientes de risco (DCC) por métodos moleculares, que melhora o fluxo diagnóstico e a eficiência da detecção.
Adenoviruses play an important role in the etiology of severe acute lower respiratory infection, especially in young children.1 Human adenoviruses (HAdV) spread rapidly in closed environments, which often cause epidemic disease in crowded communities. Furthermore, adenovirus infection is difficult to clinically distinguish from other viral or bacterial respiratory infections.2 This combination of factors indicates that improved clinical microbiological methods are necessary for detection of the acute respiratory disease caused by adenoviruses.
Prior to the advent of PCR, DFA and viral isolation were the most sensitive methods that were available for respiratory viruses detection.3,4 However, a significant number of specimens in patients with clinically compatible viral respiratory infection were incorrectly determined to be negative by DFA and viral culture, implying a failure to identify the causative virus in a significant percentage of cases.5–7 Molecular methods demonstrate greater sensitivity when compared to conventional assays for detecting adenovirus in respiratory samples.8,9
In the pediatric population HAdV is the second most common detected pathogen10 and the virus is responsible for 5%–15% of respiratory disease.10,11 Clinical samples from children often present higher viral loads when compared to adults patients.12 Brazilian studies described an incidence ranging from 3% to 7.1%, depending on the technique used.13–16 Other group at risk to acquired HAdV respiratory infection was the congenital heart disease children, but there are no published data.
In this context, the aim of the present study was to assess HAdV occurrence by two different methods (Direct Fluorescence Assay – DFA and Nested Polymerase Chain Reaction – nested PCR) in samples collected from different pediatric populations.
MethodThis cross sectional study included two different populations assessed from 2005 to 2008:
- 1.
Congenital heart disease children (CHD): during 2005, 2007, and 2008, 123 outpatients with CHD assisted in the Congenital Heart Disease Pediatric Service were included. During the year of 2006 the congenital heart disease ward underwent a structural and administrative reform, preventing sample collection. Among these patients, 49.7% were male, the mean and median age 3.9 and 2.9 years old, ranging from1 year and 6 month to 11 years old.
- 2.
Community children (CC): in 2008, 165 outpatients seen in a primary care facility by a pediatrician were included. Among these patients, 47.2% were male, the mean and median age 3.9 and 3.0 years old, ranging from 7 months to 11 years old.
For both groups, children were eligible if they presented acute respiratory infection (ARI) of probably viral etiology and preferably up to 7 days of symptoms until sample collection. In the community children group, patients were included when parents or guardians sought medical care due to acute respiratory infection. Among CHD group, the inclusion of patients was defined by a physician during a previous routine visit. The CHD population was considered at risk for viral respiratory infection complications; therefore, samples of children with more than 7 days of symptoms were collected (ranged from 1 to 60 days). Respiratory symptoms assessed were: coryza, cough, sore throat, or nasal congestion; and systemic symptoms were: fever, headache, malaise, chills, or fatigue. Written consent was obtained from all participants' legal representative before enrollment.
All nasal samples were maintained at 4 ºC and transported immediately to the laboratory. An aliquot (1 mL) was separated for molecular analysis and stored at –70 ºC. The remaining specimen volume was evaluated on the same day by direct fluorescent assay (DFA), according to previous study.17 Afterwards, one aliquot from each stored sample had DNA extracted by QIamp DNA Blood extraction kit (Qiagen, USA), according to manufacturer's instructions, follow by nested-PCR for detection of all adenovirus serotypes, as previously described.18
All studied samples were evaluated by direct immunofluorescence technique (DFA). The tests were performed using the kit “Simulfluor Respiratory Screen and Panel” (Chemicon Int., EUA), in accordance with the manufacturer's instructions.
The primers used for detection target the region of HAdV Hexon gene. The outer primer pair, hex1deg (5′-GCC SCA RTG GKC WTA CAT GCA CAT C-3′) and hex2deg (5′-CAG CAC SCC ICG RAT GTC AAA-3'), resulted in a 301 bp product. The nested primer pair, nehex3deg (5′-GCC CGY GCM ACI GAI ACS TAC TTC-3′) and nehex4deg (5′-CCY ACR GCC AGI GTR WAI CGM RCY TTG TA-3′), produced an amplicon of 171 bp. For the first amplification reaction, 5 µL of extracted DNA were placed in a tube with a reaction mixture consisting of 2.5 µL of 10× buffer (200mM Tris-HCl, pH 8.4, 500mM KCl), 3.5 µM of MgCl2, 0.5 µM of primers Hex1deg and Hex2deg, 1µL of dNTPs mixture containing 20 mM of each nucleotide, 2,5U of Platinum® TaqDNA Polymerase (Invitrogen, Brazil) and autoclaved MilliQ water to a final volume of 25 µL. For the second amplification reaction, 2 µL of first reaction amplicon were placed in a tube with a reaction mixture consisting of 2.5 µL of 10× buffer (200 mM Tris-HCl, pH 8.4, 500 mM KCl), 3.5 µM of MgCl2, 0.5 µM of primers Nehex3deg and Nehex4deg, 1µL of dNTPs mixture containing 20 mM of each nucleotide, 2,5U of Platinum® TaqDNA Polymerase (Invitrogen, Brazil) and autoclaved MilliQ water to a final volume of 25 µL. Positive controls (Adenovirus Serotype 3) and redundant negative control (autoclaved MilliQ water) were included in each series. Positive results were considered when amplicon were visualized after 2% agarose gel electrophoresis. The sensibility of the reaction were standardized and showed a detection limit of 10−4 TCID50/mL.
Association between different categorical variables (response variable: presence or absence of HAdV; independent variable: community children and congenital heart disease children) was tested using the chi-square test and univariate Odds Ratio (SPSS, version 11.5). p<0.05 was considered to be statistically significant. To compare used methods, the Kappa test was conducted (R software version 2.11.1) and the interpretation of the coefficients was performed using the classification of Altman.19
The present study was approved by the Institutional Review Board of São Paulo Hospital and the Federal University of São Paulo (1654/09).
ResultsSamples from 290 symptomatic patients were analyzed (48.6% (141/290) females and 51.4% (149/290) males), being 56.9% (165/290) from the community and 43.1% (125/290) from CHD children. The time between onset of symptoms and sample collection ranged from 1 to 60 days.
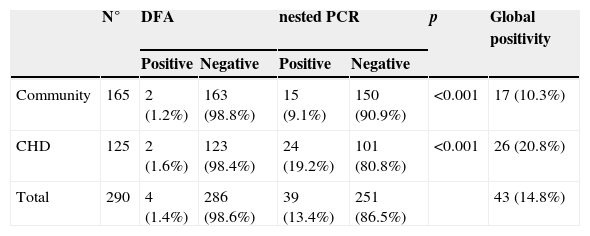
Among the 290 samples included during the study period, 41 (14.1%) were positive for at least one test: 17/165 (10.3%) children from community, 26/125 (20.8%) CHD children. Table 1 show the differential detection between assays according to different studied populations. Overall, 4/290 (0.7% of samples were DFA positive and 39/290 (13.40%) were nested-PCR positive. Nested-PCR presented a higher detection, statistically significant, among all studied populations when compared to DFA (p<0.001). Comparing the children groups by molecular methods, CHD presented higher detection rate (p=0.02).
HAdV detection by DFA and nested-PCR among different studied populations.
| N° | DFA | nested PCR | p | Global positivity | |||
|---|---|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | ||||
| Community | 165 | 2 (1.2%) | 163 (98.8%) | 15 (9.1%) | 150 (90.9%) | <0.001 | 17 (10.3%) |
| CHD | 125 | 2 (1.6%) | 123 (98.4%) | 24 (19.2%) | 101 (80.8%) | <0.001 | 26 (20.8%) |
| Total | 290 | 4 (1.4%) | 286 (98.6%) | 39 (13.4%) | 251 (86.5%) | 43 (14.8%) | |
CHD, children with congenital heart disease; DFA, direct fluorescence assay; PCR, Polymerase Chain Reaction.
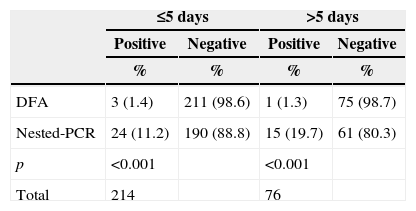
Analysis of the length of time between symptoms onset and the sample collection day showed a statistical difference between DFA and nested-PCR (Table 2). Among community children, the mean ± standard deviation, median and range since symptoms onset were: 3.2±2.72; 3; 1-20 days; and among CHD children the mean ± standard deviation, median and range since symptoms onset were: 6.5±7.1; 5; 1-60 days. Nested-PCR compared to DFA detected a higher number of cases both for samples collected within 5 days of symptoms onset, and >5 days of symptoms onset.
Time of symptoms onset according to evaluated test.
| ≤5 days | >5 days | |||
|---|---|---|---|---|
| Positive | Negative | Positive | Negative | |
| % | % | % | % | |
| DFA | 3 (1.4) | 211 (98.6) | 1 (1.3) | 75 (98.7) |
| Nested-PCR | 24 (11.2) | 190 (88.8) | 15 (19.7) | 61 (80.3) |
| p | <0.001 | <0.001 | ||
| Total | 214 | 76 | ||
DFA, direct fluorescence assay; PCR, Polymerase Chain Reaction.
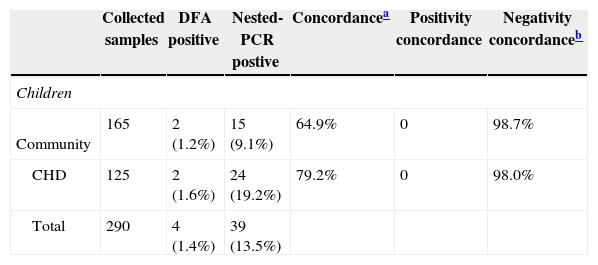
Table 3 shows the agreement between the evaluated tests. The general agreement was low, mainly among community children. The concordance regarding negative results was high for both studied populations.
Agreement assessment between the tests.
| Collected samples | DFA positive | Nested-PCR postive | Concordancea | Positivity concordance | Negativity concordanceb | |
|---|---|---|---|---|---|---|
| Children | ||||||
| Community | 165 | 2 (1.2%) | 15 (9.1%) | 64.9% | 0 | 98.7% |
| CHD | 125 | 2 (1.6%) | 24 (19.2%) | 79.2% | 0 | 98.0% |
| Total | 290 | 4 (1.4%) | 39 (13.5%) | |||
CHD, children with congenital heart disease; PCR, Polymerase Chain Reaction.
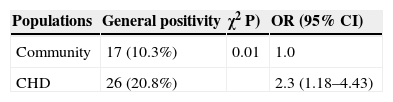
Table 4 shows the result of the univariate Odds Ratio analysis of HAdV occurrence among different studied populations. The response variable was the presence or absence of HAdV, and the independent variable was the studied group: community children and CHD children. CHD children presented approximately two times more chance to acquired HAdV infection when compared to community children.
DiscussionAmong community children, the adenovirus detection rate was similar to that described in the national literature, with rates ranging from 6% to 7.1%.13–16 A recent Brazilian study evaluating 1,121 samples collected from hospitalized infants found adenovirus occurrence in 15.8%, which corroborates the findings of the present study.10 International studies report HAdV occurrence ranging from 2%-3.8% among pediatric populations, which is slightly lower when compared to our results.20–22 Studies with CHD children demonstrate respiratory viruses circulation, with relative high occurrence rates, showing that these agents must be considered in clinical and diagnostic practice.22 The present study is the first to describe HAdV respiratory infection in Brazilian children with congenital heart disease. The HAdV detection rate in CHD children was higher (20.8%) when compared to community children (10.3%). It is noteworthy that, among CHD children sample collection was accomplished during routine consultation. If the patient presented respiratory infection during the scheduled appointment, the sample was collected. Therefore the rate found could be underestimated, because probable cases may have been lost due to lack of active demand from patients. One hypothesis for the higher detection can be related to comorbidities presence among this children.23
When evaluating the two tests used in this study some important questions for both clinicians and hospital administrators are raised: Which is the best test? Up to which day is ideal to collect the sample? Is there a need to repeat the test? Which is the ideal number of samples for laboratory routine flow? What is the impact of negative and positive tests for high-risk patients? Some of these aspects could be evaluated in the present study.
The analysis of the association between patient´s symptom onset and laboratorial test results showed that the molecular method was always more sensible than the DFA, and this difference was more evident after the fifth day of symptoms (p<0.001). Lower detection by DFA was related to low viral load in respiratory samples.24 The higher detection rates among patients with more than five days of symptom were expected due to higher nested-PCR sensibility.25,26 The success of DFA test depend on some key factors as good sample collection and experienced laboratory staff,10 while nested-PCR, based on nucleic acid amplification, can detect a very low number of viral particles.10,26
Univariate analysis showed that CHD children have a higher risk of acquiring HAdV (Odds Ratio: 2.29), when compared to community children. Utokaparch (2011)27 described that pediatric patients with lower respiratory infection presented higher viral load when compared to patients with upper respiratory infection. Echavarria et al.28 described that children are 2 to 3.5 times more likely to become infected by HAdV than adults. These data corroborates the results of the present study.
One of the study limitations was the lack of another respiratory viruses analysis. Among HAdV negative samples, others virus could be present. As a result of the high sensitivity of the molecular methods, viral infections could be detected weeks after the symptoms resolution29,30 but, in the present study, the included samples were collected from children presenting acute respiratory infection, decreasing the chance of a past infection detection. Another study limitation was the lack of sample collection during 2006; however the main idea of the study was to understand the occurrence of the virus in the community (community children), and in a population with at high risk of complications after a viral respiratory infection (CHD children). Some studies described an occurrence of adenovirus during different years, and throughout the year,31,32 with similar occurrence patterns; therefore the collection of samples during different years apparently did not compromise the purpose of the study.
Three parameters can indicate the validity of the diagnostic test choice, the cost per sample, the turnaround time (time to provide the result) and the test reliability. In a study published in 2009 Mahony et al.33 compared the cost of DFA versus molecular technique for diagnosing respiratory virus infection, and they described that molecular test was slightly more expensive but, at the same time, was highly cost-effective due to reduction in hospitalization time. Different studies described the turnaround time of DFA and PCR, with both tests presenting the ability to provide results in one day.34,35 Gharabaghi et al.34 compared the sensitivity and specificity of DFA and commercial molecular assay. This study showed lower sensitivity of DFA test in comparison to molecular testing for all surveyed virus, with adenovirus presenting the lowest sensitivity (38.1%)34
We suggest that routine surveillance should be performed in high-risk patients by molecular methods, improving diagnostic flow and efficiency. Our results also demonstrated that in samples with more than five days of symptom onset nested-PCR might be the best option.
FundingCNPq study grant (DP).
Conflicts of interestThe authors declare no conflicts of interest.