The increasing presence of antibiotic-resistant bacteria in urban agriculture poses a significant public health risk. This study investigated the prevalence and virulence of methicillin-resistant Staphylococcus aureus (MRSA) in market garden environments in Bobo-Dioulasso, Burkina Faso. A total of 135 environmental samples (lettuce, irrigation water, and organic manure) were analyzed for MRSA presence using microbiological isolation, antimicrobial susceptibility testing, and molecular characterization. Among 55 Staphylococcus isolates, 16 (32.7%) were confirmed as MRSA, with PVL detected in 18.4% and TSST-1 in 6.1%. High resistance rates to oxacillin (96.4%) and fusidic acid (69.1%) highlight the need for targeted antimicrobial stewardship. Statistical analyses were conducted using R (version 2.10.0), applying Chi-square, ANOVA, Kruskal–Wallis, and logistic regression tests to assess variations in antibiotic resistance and predictive factors in environmental samples, with significance set at p<0.05. These findings underscore the necessity for enhanced microbial surveillance, improved hygiene protocols, and policy interventions to mitigate foodborne risks associated with urban agriculture.
La creciente presencia de bacterias resistentes a los antibióticos en la agricultura urbana representa un riesgo importante para la salud pública. Este estudio investigó la prevalencia y la virulencia de Staphylococcus aureus resistente a la meticilina (MRSA) en entornos hortícolas de Bobo-Dioulasso, Burkina Faso. Se analizaron 135 muestras ambientales (lechuga, agua de riego y estiércol orgánico) para detectar MRSA mediante aislamiento microbiológico, pruebas de sensibilidad antimicrobiana y caracterización molecular. Los análisis estadísticos se realizaron con R (versión 2.10.0), aplicando pruebas de Chi-cuadrado, ANOVA, Kruskal-Wallis y regresión logística para evaluar las variaciones en la resistencia a antibióticos y los factores predictivos en las muestras ambientales, con un nivel de significancia establecido en p<0.05. Entre los 55 aislamientos de Staphylococcus obtenidos, 16 (32,7%) fueron confirmados como MRSA, con detección de leucocidina de Panton-Valentine (PVL) en el 18,4% y toxina-1 del síndrome de shock tóxico (TSST-1) en el 6,1%. Las altas tasas de resistencia a oxacilina (96,4%) y ácido fusídico (69,1%) destacan la necesidad de una gestión antimicrobiana dirigida. Nuestros hallazgos subrayan la necesidad de una vigilancia microbiana reforzada, de protocolos de higiene mejorados y de intervenciones políticas para mitigar los riesgos alimentarios asociados con la agricultura urbana.
Urban agriculture plays a crucial role in food security and economic stability in rapidly growing cities. However, it also poses significant food safety concerns due to microbial contamination, particularly in fresh produce such as lettuce7,8. Methicillin-resistant Staphylococcus aureus (MRSA), a multi-drug-resistant pathogen, poses a notable risk in foodborne transmission, yet its presence in urban agricultural environments remains underexplored in low-resource settings9. Leafy vegetables, especially lettuce, are prone to bacterial contamination due to direct exposure to irrigation water and organic manure during cultivation5. These matrices serve as potential reservoirs for pathogen transfer, reinforcing the need for targeted microbiological surveillance6. The presence of virulence-associated genes such as Panton-Valentine leukocidin (PVL) and toxic shock syndrome toxin-1 (TSST-1) in MRSA strains further exacerbates public health concerns, as these toxins contribute to severe infections in humans12. While MRSA is extensively studied in clinical settings, data on its prevalence, resistance patterns, and virulence factors in market garden produce remain scarce2. Integrating a One Health approach to microbial surveillance is critical to understanding the transmission dynamics of antibiotic-resistant bacteria in food systems13. This study aimed to investigate the presence of methicillin-resistant S. aureus (MRSA) in lettuce, irrigation water, and organic manure across three urban market garden sites in Bobo-Dioulasso, Burkina Faso. Specifically, it sought to determine the prevalence of S. aureus strains, analyze their antibiotic resistance profiles, detect virulence genes (PVL, TSST-1, enterotoxins), and examine environmental factors that may influence their dissemination, within an integrated One Health perspective.
Materials and methodsStudy frameworkThis study was conducted at three market garden sites in Bobo-Dioulasso, Burkina Faso. Microbiological analyses were performed at the Microbiology Laboratory of the Food Technology Department (DTA) at IRSAT/C NRST, Western Regional Directorate, Bobo-Dioulasso, while molecular analyses were conducted at the Molecular Biology and Genetics Laboratory (LaBioGEN), UFR/SVT, Joseph KI-ZERBO University, Ouagadougou.
Study type and durationThis descriptive cross-sectional study, involving prospective sampling, was conducted over a 10-month period, between March 1 to December 31, 2023.
Sampling strategySampling was conducted at three market gardening sites in Bobo-Dioulasso: Kuinima, Kodeni and Sakabi. These three sampling sites are located on the outskirts of Bobo-Dioulasso and exhibit distinct yet complementary agro-environmental characteristics. Kuinima is a peri-urban zone adjacent to a protected forest area, where intensive market gardening relies heavily on untreated irrigation water and organic manure, exposing the site to significant anthropogenic pressure. Kodeni is a densely populated and rapidly expanding neighborhood that combines small-scale farming and poultry rearing, with mixed use of organic and chemical fertilizers and strong rural–urban interactions. Sakabi, in contrast, is a semi-rural site with limited urbanization, where extensive vegetable farming depends on natural resources such as runoff water and traditional wells. While all three sites engage in agricultural activities, their differences in human density, irrigation practices, and proximity to pollution sources may influence the microbiological profiles observed.
A total of 135 samples, comprising lettuce (n=45), irrigation water (n=45), and organic manure (n=45), were randomly collected from farmers. At each site, 45 samples including 15 lettuce samples, 15 irrigation water samples and 15 manure samples were collected. Samples were randomly collected from five (5) gardeners at each site, at a rate of three (3) samples (i.e., lettuce (n=1), irrigation water (n=1), organic manure (n=1)) per gardener and per week for three (3) weeks. Lettuce and manure samples were placed and securely sealed in sterile plastic bags while water samples were collected in sterile labeled jars. All samples were transported at 4°C in insulated containers with ice packs to the Microbiology Laboratory of the Food Technology Department (DTA/IRSAT) for further analysis.
Isolation and identification of S. aureusThe Baird-Parker medium (Himedia, India), enriched with egg yolk and potassium tellurite (Himedia, India) was used for the selective isolation of Staphylococcus species following ISO 6888-3 standards. Sample preparation involved mixing 10g of each sample with 90ml of sterile diluent in a sterile stomacher bag, following ISO6887-1 (1999) guidelines. The mixture was homogenized for 2min using a stomacher to obtain a stock suspension (1:10 dilution). The prepared samples were then inoculated onto Baird-Parker yellow agar supplemented with potassium tellurite. The inoculated Petri dishes were placed in an incubator, previously sanitized with 65% alcohol, and set to 37±1.0°C for 24±2h. All procedures were carried out aseptically on a bench thoroughly cleaned with 70% alcohol. After the stipulated incubation time, growth resulted in the characteristic S. aureus colonies surrounded by a clear halo. Single purified S. aureus colonies were then cultured onto Brain Heart Infusion media (Himedia, India) for 24h at 37°C. A coagulase test was performed using lyophilized rabbit plasma (Himedia, India) to differentiate S. aureus from other staphylococcal species. Five colonies were subcultured in 5ml of Oxoid Brain Heart Infusion broth and incubated at 37°C for 24h. After incubation, 0.5ml of the culture was mixed with 0.5ml of rabbit plasma, shaken well, and incubated at 37°C for 6–24h. The formation of a 2/3 coagulum in the tube was considered a positive coagulase reaction. Confirmed coagulase-positive colonies were subsequently subcultured onto Brain Heart Infusion broth supplemented with 15% glycerol and stored at −80°C for future analyses.
Antibiotic susceptibility testingThe disk diffusion method13 was performed following the guidelines of the French Society for Microbiology3. Confirmed coagulase-positive colonies were subcultured on Mueller-Hinton (MH) agar and incubated at 37°C for 18–24h. A bacterial suspension was prepared by emulsifying 2–3 colonies in 10ml of 0.9% saline (NaCl) and then swabbed evenly onto MH agar plates. Antibiotic disks were placed within 15min using sterile forceps. The tested antibiotics include: penicillin G (10μg/disk), amikacin (30μg/disk), gentamicin (10μg/disk), ciprofloxacin (5μg/disk), tobramycin (10μg/disk), erythromycin (15μg/disk), fusidic acid (10μg/disk), cefoxitin (30μg/disk), oxacillin (1μg/disk), amoxiclav (10.5μg/disk), and netilmicin (30μg/disk) (Oxoid, UK). Reference strains (S. aureus ATCC 43300 and ATC 29213) were used for quality control. Following 24h of incubation, inhibition zone diameters were measured using calipers, and isolates were classified as susceptible or resistant based on CA-SFM (2020) guidelines. Three Petri dishes were used per sample: one for cefoxitin, while the remaining two dishes contained five antibiotic disks each.
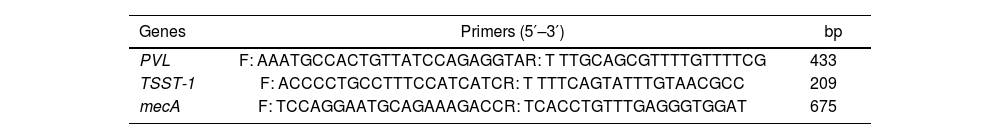
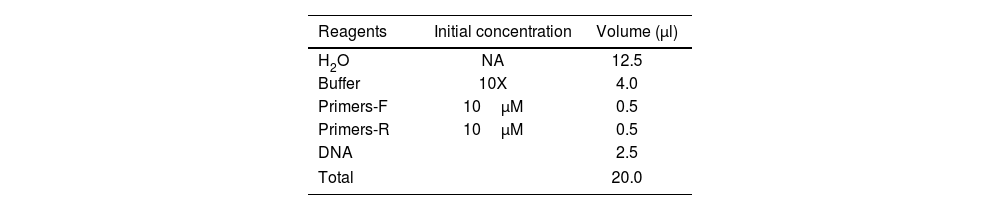
Molecular characterization of S. aureus strainsPolymerase chain reaction (PCR) was performed to amplify and detect specific DNA fragments. DNA extraction was performed using the thermolysis method. Two or three (2–3) isolated colonies were suspended in 300μl of sterile distilled water. The mixture was heat-treated at 100°C for 15min, followed by centrifugation at 12000rpm for 15min. The resulting supernatant (DNA matrix) was stored at −20°C for future analyses. S. aureus isolates underwent 16S-23S genomic analysis using primers G1 (5′-GAAGTCGTAACAAGG-3′) and L1 (5′-CAAGGCATCCACCGT-3′). Screening for PVL, TSST-1 and mecA genes was performed using specific primers (Table 1). Amplification was conducted using a thermal cycler, employing universal primers in a 20μl reaction volume containing the DNA sample (Table 2). A specific PCR program was designed for each target gene. The PCR protocol for DNA isolate characterization consisted of an initial denaturation at 94°C for 2min. The amplification protocol followed this sequence: initial denaturation at 94°C for 3min, 35 cycles, each comprising: denaturation at 94°C for 1min, annealing at 50°C (PVL and TSST-1), and 55°C (mecA) for 1min, extension at 72°C for 1min and final extension at 72°C for 10min. Reference strain S. aureus ATCC 43300 and PCR water were used for positive and negative quality control respectively.
A 1.5% agarose gel was prepared by dissolving agarose powder in 100ml of 1X TAE buffer (Tris–Acetate–EDTA). The mixture was heated in a microwave for approximately 5min until complete dissolution, yielding a clear and homogenous solution. After cooling to ∼50°C, 0.15μl of ethidium bromide was added. The prepared gel was poured into a casting tray with a comb, forming sample wells for PCR amplicon deposition. The gel was placed in an electrophoresis chamber containing electrophoresis buffer. Each well was loaded with approximately 8μl of amplicon, following a predefined layout. Additionally, the first well in each row was loaded with 8μl of a 100× molecular weight marker, which contained DNA fragments of known sizes serving as reference points. Electrophoresis was performed in 0.5X TAE buffer at a voltage of 120V for 30min. Following separation, DNA fragment sizes were determined using a 100bp standard molecular weight marker while bands were visualized under UV light using a Vilber E-BOX imaging system.
MRSA confirmationCefoxitin disks (30μg) were utilized to identify methicillin-resistant isolates, with an inhibition zone diameter <27mm indicating MRSA. S. aureus ATCC 25923 serves as the quality control strain. The mecA gene, known to confer methicillin resistance in S. aureus (MRSA), was detected by PCR using primers as described previously.
Statistical analysisStudy data was systematically collected using predefined survey forms, followed by entry and processing utilizing Microsoft Office 2010 Excel. Statistical analyses were conducted using R (version 2.10.0) to ensure a robust validation of findings. Chi-square test (R) was used to compare the prevalence of resistant strains between matrices (p=0.02). ANOVA test (R) was applied to determine variations in resistance across sites (p=0.03). Kruskal–Wallis test (R) was used as a non-parametric alternative when data did not meet normality assumptions. Logistic regression (R) was used to identify predictive factors for MRSA presence, highlighting the influence of lettuce and organic manure (p<0.05). Data were cleaned and structured in Excel CSV format before being imported into R. Normality was verified using Shapiro–Wilk and Kolmogorov–Smirnov tests. Results were reported with 95% confidence intervals (CI) and statistical significance set at p<0.05.
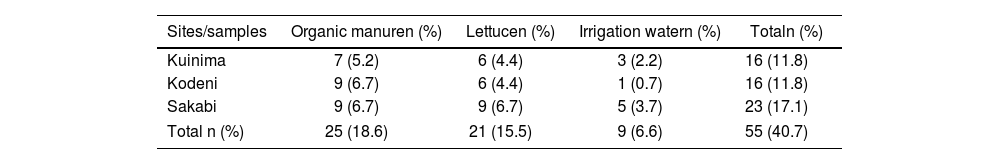
ResultsPrevalence of Staphylococcus strains isolated from three market garden sites in Bobo-DioulassoTable 3 provides a breakdown of the prevalence of Staphylococcus strains from different sample types and collection sites. A total of 135 samples were collected from three market garden sites in Bobo-Dioulasso: Kuinima, Kodeni, and Sakabi. The samples consisted of organic manure (n=45), lettuce (n=45), and irrigation water (n=45). Among these, 55 Staphylococcus strains (40.7%) were successfully isolated and confirmed positive by the coagulase test. The distribution by collection site included Sakabi (23 strains, 17.1%), Kodeni (16 strains, 11.8%), Kuinima (16 strains, 11.8%). By sample type, organic manure (25 strains, 18.6%), lettuce (21 strains, 15.5%), irrigation water (9 strains, 6.6%). The Chi-square test (p=0.02) shows a significant difference in strain distribution across matrices, with organic manure showing the highest concentration.
Prevalence of Staphylococcus aureus strains isolated from market gardening sites in Bobo-Dioulasso.
| Sites/samples | Organic manuren (%) | Lettucen (%) | Irrigation watern (%) | Totaln (%) |
|---|---|---|---|---|
| Kuinima | 7 (5.2) | 6 (4.4) | 3 (2.2) | 16 (11.8) |
| Kodeni | 9 (6.7) | 6 (4.4) | 1 (0.7) | 16 (11.8) |
| Sakabi | 9 (6.7) | 9 (6.7) | 5 (3.7) | 23 (17.1) |
| Total n (%) | 25 (18.6) | 21 (15.5) | 9 (6.6) | 55 (40.7) |
n: number of isolates; %: percentage.
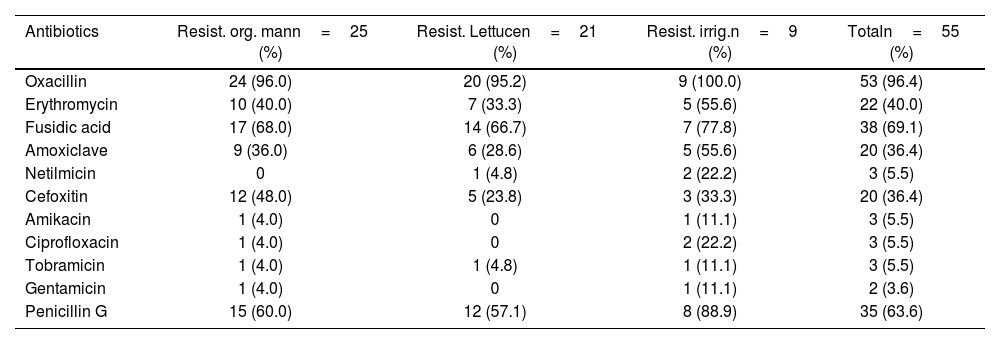
Antibiotic susceptibility testing revealed high resistance levels among coagulase-positive Staphylococcus isolates. The highest resistance rates were observed against oxacillin (96.4% resistance), confirms the presence of MRSA; fusidic acid (69.1% resistance); penicillin G (63.6% resistance); gentamicin, ciprofloxacin, tobramycin, amikacin, and netilmicin (low resistance (≤5.5%) (Table 4). ANOVA test (p=0.03) shows significant variation in antibiotic resistance across sites and sample matrices.
Resistance profile of isolated Staphylococcus aureus strains.
| Antibiotics | Resist. org. mann=25 (%) | Resist. Lettucen=21 (%) | Resist. irrig.n=9 (%) | Totaln=55 (%) |
|---|---|---|---|---|
| Oxacillin | 24 (96.0) | 20 (95.2) | 9 (100.0) | 53 (96.4) |
| Erythromycin | 10 (40.0) | 7 (33.3) | 5 (55.6) | 22 (40.0) |
| Fusidic acid | 17 (68.0) | 14 (66.7) | 7 (77.8) | 38 (69.1) |
| Amoxiclave | 9 (36.0) | 6 (28.6) | 5 (55.6) | 20 (36.4) |
| Netilmicin | 0 | 1 (4.8) | 2 (22.2) | 3 (5.5) |
| Cefoxitin | 12 (48.0) | 5 (23.8) | 3 (33.3) | 20 (36.4) |
| Amikacin | 1 (4.0) | 0 | 1 (11.1) | 3 (5.5) |
| Ciprofloxacin | 1 (4.0) | 0 | 2 (22.2) | 3 (5.5) |
| Tobramicin | 1 (4.0) | 1 (4.8) | 1 (11.1) | 3 (5.5) |
| Gentamicin | 1 (4.0) | 0 | 1 (11.1) | 2 (3.6) |
| Penicillin G | 15 (60.0) | 12 (57.1) | 8 (88.9) | 35 (63.6) |
Resist. org. man: resistance of strains isolated from organic manure; Resist. Lettuce: resistance of strains isolated from lettuce; Resist. irrig.: resistance of strains isolated from irrigation water; n: number of isolates; %: percentage.
Strains isolated from irrigation water displayed the highest resistance levels, followed by organic manure, with lettuce isolates showing relatively lower resistance (Table 4).
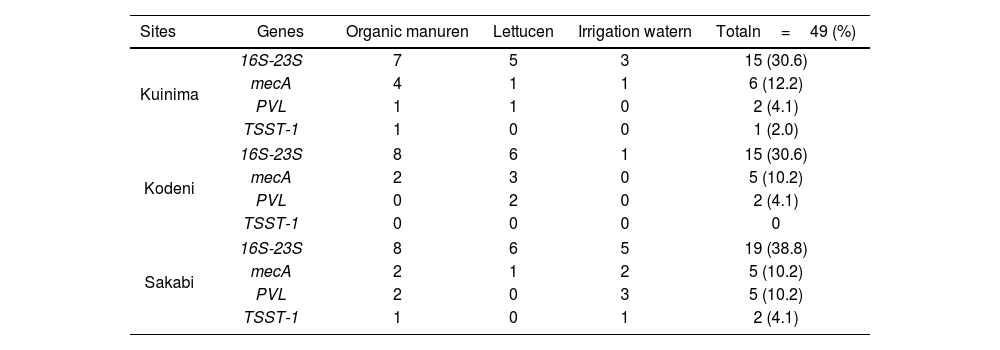
Molecular characterizationAmong the 55 coagulase-positive Staphylococcus strains, 49 (89.1%) were confirmed as S. aureus through molecular analysis. Sixteen (16) strains (32.6%) were identified as methicillin-resistant S. aureus (MRSA), with distribution across all three sites. The mecA gene was detected in multiple isolates, confirming methicillin resistance. This study demonstrated the presence of virulence genes (PVL gene: 9 strains, 18.4%), TSST-1 gene: 3 strains, 5.1%) (Table 5). Logistic regression analysis (p<0.05 for lettuce and organic manure) shows that these matrices significantly influence MRSA presence (Table 5).
Distribution of virulence and mecA genes according to sample type and collection site.
| Sites | Genes | Organic manuren | Lettucen | Irrigation watern | Totaln=49 (%) |
|---|---|---|---|---|---|
| Kuinima | 16S-23S | 7 | 5 | 3 | 15 (30.6) |
| mecA | 4 | 1 | 1 | 6 (12.2) | |
| PVL | 1 | 1 | 0 | 2 (4.1) | |
| TSST-1 | 1 | 0 | 0 | 1 (2.0) | |
| Kodeni | 16S-23S | 8 | 6 | 1 | 15 (30.6) |
| mecA | 2 | 3 | 0 | 5 (10.2) | |
| PVL | 0 | 2 | 0 | 2 (4.1) | |
| TSST-1 | 0 | 0 | 0 | 0 | |
| Sakabi | 16S-23S | 8 | 6 | 5 | 19 (38.8) |
| mecA | 2 | 1 | 2 | 5 (10.2) | |
| PVL | 2 | 0 | 3 | 5 (10.2) | |
| TSST-1 | 1 | 0 | 1 | 2 (4.1) | |
PVL: Panton-Valentine leukocidin; TSST-1: toxic shock syndrome toxin-1; n: number of isolates; %: percentage.
Food contamination remains a major public health concern, particularly when such foods are intended for human consumption. The microbiological analysis of the samples examined in our study revealed sub-optimal quality, with 55 out of 135 samples testing positive for Staphylococcus, of which 49 isolates were MRSA. Our findings indicate that lettuce, irrigation water, and organic manure harbor a diverse range of microorganisms, including S. aureus. These results highlight the critical role of irrigation water and organic soil amendments in the contamination of produce such as lettuce in market gardens.
Notably, irrigation water sourced from stagnant pools, drainage canals, and waste disposal wells appears to be a significant reservoir of S. aureus contamination20. Studies have also shown that stagnant water and drainage canals are frequently contaminated with fecal matter, particularly from animal manure used as fertilizer14. Moreover, these water sources are vulnerable to contamination from human activities, further intensifying public health risks. The systematic recovery of resistant S. aureus in market environments suggests active circulation of resistance genes, likely due to agricultural practices that heighten selective pressure for resistance. This highlights the broader role of environmental factors in the spread of antimicrobial resistance, reinforcing the importance of strengthened surveillance and intervention measures.
Coagulase-positive S. aureus strains were identified in three market gardens (Sakabi, Kodeni, and Kuinima) with prevalence rates of 17.1% (23 strains), 11.8% (16 strains), and 11.8% (16 strains) respectively. While slight variations were present across sites, these findings suggest that market gardeners commonly engage in similar agricultural practices, including the use of wastewater for irrigation and the application of uncomposted animal feces, both of which increase the risk of bacterial contamination. This observation aligns with previous reports, as organic manure used for soil fertilization infrequently undergoes the recommended composting processes in resource-poor settings, rendering it highly susceptible to microbial contamination.
Several studies have demonstrated that animal feces are key reservoirs for bacterial pathogens in agricultural settings10. Due to limited access to mineral fertilizers, many farmers rely on animal excrement as a cost-effective and environmentally sustainable alternative. However, Kagambèga et al.10, found that animal feces (including those from cattle, poultry, pigs, and hedgehogs) harbor diverse bacterial pathogens, further emphasizing their role in pathogen dissemination. The lack of proper manure composting presents a significant public health hazard, increasing the likelihood of contamination of fresh produce and water sources.
Multiple studies have identified poor hygiene practices among farmers as playing a major role in S. aureus transmission, given its propensity to colonize the skin and mucous membranes, particularly the hands and nasal passages20,1,15. Furthermore, Enterobacteriaceae and Staphylococcus species can persist on contaminated surfaces (including bedding, clothing, and work equipment) exacerbating the risk of environmental dissemination. Extensive handling of vegetables significantly increases contamination risks associated with S. aureus, as demonstrated in prior research19.
The detection of MRSA strains in irrigation water, soil, and manure in this study suggests that urban market gardening sites may serve as environmental reservoirs for resistant bacteria. This poses an increased risk of exposure to farmers, consumers, and surrounding communities, potentially leading to infections that are difficult to treat.
The high prevalence of multidrug resistance (most S. aureus isolates exhibiting resistance to at least three antibiotics) underscores a serious public health concern. The implementation of robust intervention strategies is essential to monitor and mitigate the transmission of resistant strains, as their dissemination is likely to intensify in the absence of stringent enforcement of agricultural hygiene standards.
This study revealed high resistance rates to oxacillin, fusidic acid, and penicillin G, which is consistent with findings from previous studies21,18,22,17. The widespread use of these antibiotics in both animal husbandry and human medicine, coupled with their unregulated availability, has contributed to the increasing prevalence of resistant strains over time. Environmental and agricultural reservoirs of resistant bacteria pose a major challenge in controlling infections and emphasize the urgency of enhanced antibiotic stewardship strategies. Vegetables grown in these environments may be contaminated with resistant strains, especially in the absence of proper post-harvest hygiene practices. This situation raises concerns about food safety and consumer health, particularly among vulnerable populations.
S. aureus harbors multiple virulent determinants contributing to its pathogenicity, with agricultural products and poor farming practices serving as potential catalysts for its environmental dissemination. Among its virulence factors, toxins play a major role23. In our study, the PVL gene was detected in 18.4% of isolates, while the TSST-1 gene was identified in 6.1%. These findings are consistent with previous research demonstrating a high prevalence of PVL and TSST-1 positive S. aureus isolates among pigs and pig farm workers in Nigeria and South Africa11,16.
Furthermore, five isolates carried both the mecA and PVL genes, one harbored both mecA and TSST-1, and two contained both PVL and TSST-1. The presence of MRSA in food and environmental sources, combined with its resistance profile, underscores the need for urgent intervention. Given that lettuce is commonly consumed raw, the risk of transmission to humans is particularly concerning. According to the World Health Organization (WHO), antimicrobial resistance ranks among the top 10 global public health threats to humanity4.
The observed differences in antimicrobial resistance and virulence gene profiles between S. aureus isolates from irrigation water and those from organic manure may reflect distinct environmental selection pressures and microbial dynamics. Isolates recovered from irrigation water exhibited greater resistance, potentially due to the continuous exposure to low concentrations of antibiotics and other contaminants originating from urban runoff, domestic wastewater, or agricultural effluents. Such environments can promote the survival and proliferation of resistant strains through selective pressure and horizontal gene transfer. In contrast, isolates from organic manure showed a higher prevalence of virulence-associated genes and toxins, which may be attributed to the microbial composition of animal feces and the presence of commensal or pathogenic strains with enhanced genetic potential. The nutrient-rich and anaerobic conditions of manure may favor the persistence of toxigenic S. aureus strains, particularly those carrying PVL, TSST-1, or enterotoxin genes. These findings underscore the importance of environmental reservoirs in shaping the resistance and pathogenicity profiles of S. aureus within urban agricultural systems.
Our study highlights the urgent need for coordinated action among public health professionals, sanitation officials, farmers, and municipal policymakers to implement stringent and effective food safety measures. Additionally, this study provides essential local epidemiological data, contributing to efforts to combat antimicrobial resistance within a One Health perspective. Strengthening microbiological surveillance, enhancing agricultural hygiene practices, and implementing stricter antibiotic regulations will be critical in mitigating the risks associated with the persistence and transmission of resistant S. aureus strains.
Study limitationsRestricted geographical samplingThis study was conducted at three urban market gardening sites in Bobo-Dioulasso, which limits the extrapolation of the findings to other regions or the national level. Environmental conditions and agricultural practices may differ significantly between sites, potentially influencing the presence and distribution of MRSA.
Sample size constraintsThe sample size (covering water, soil, and manure) may be insufficient to detect strong trends or statistically significant associations. This limitation may affect the robustness of the conclusions regarding prevalence and resistance profiles.
Lack of human and animal dataThe One Health approach would have been strengthened by including samples from humans (e.g., farmers, consumers) or animals (e.g., livestock, domestic pets). Such data could have helped establish clearer epidemiological links between environmental MRSA and potential reservoirs or transmission pathways.
These findings underscore the importance of implementing integrated antimicrobial resistance (AMR) surveillance systems within urban agricultural environments. Such systems should be designed under a One Health perspective, requiring coordinated efforts across human, animal, and environmental health sectors. In addition, farmers and fresh produce vendors must be made aware of the risks associated with AMR and trained in safer agricultural and hygiene practices. Community-level awareness campaigns could play a critical role in reducing risky behaviors and promoting public health. Furthermore, the data generated by this study may inform national AMR strategies by highlighting urban agriculture as a priority area for intervention. These insights could also contribute to Burkina Faso's engagement with the WHO Global Action Plan on AMR, reinforcing the need for context-specific policies and multisectoral collaboration.
ConclusionThis study, which aimed to characterize virulence genes associated with toxin production in methicillin-resistant S. aureus (MRSA) strains isolated from three market garden sites in Bobo-Dioulasso, yielded significant findings. The results confirm that S. aureus contamination is widespread in these agricultural environments, which could largely be due to poor farming practices. Among the isolated strains, MRSA exhibited high resistance to commonly used antibiotics, highlighting a serious public health concern. Additionally, the detection of virulence genes encoding PVL and TSST-1 toxins that suggest high pathogenic potential, along with the presence of genes associated with multidrug resistance, may further heighten the public health risk. The contamination of lettuce plants and its surrounding environment pose a substantial risk for foodborne illnesses, which can potentially lead to severe and difficult-to-treat infections. There is urgent need for advocacy and capacity-building initiatives among market gardeners, to promote optimal agricultural practices that minimize contamination risks. Likewise, consumer awareness campaigns should emphasize the importance of thoroughly washing fresh produce before consumption, ensuring good safety and minimizing health hazards.
CRediT authorship contribution statementDSK: Writing original draft, Validation, Methodology, Investigation, Formal analysis, Data curation.
NSS: Writing original draft, Validation, Supervision, Methodology, Formal analysis, Conceptualization.
MAKC: Writing – review & editing, Validation, Supervision, Methodology, Formal analysis, Conceptualization.
YB: Writing – review & editing, Validation, Resources, Methodology, Investigation.
AT and NMA: Writing – review & editing, Validation, Resources, Methodology, Investigation.
AM-D: Writing – original draft, Validation, Supervision, Project administration, Methodology, Data curation, Conceptualization.
Ethics approvalNot applicable.
Consent to publishAll authors have read and approved the final version of the manuscript for publication.
Declaration of generative AI and AI-assisted technologies in the writing processDuring the preparation of this work, the author(s) used Microsoft Copilot to assist with improving the clarity, academic style, and linguistic precision of the manuscript. After using this tool, the author(s) carefully reviewed and edited all content, and take(s) full responsibility for the final version of the publication.
FundingNone declared.
Declaration of competing interestThe authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
We sincerely would like to thank the DTA for their confirmation and serotyping of S. aureus isolates used in this study. We declare no conflicts of interest.