The interpretation of medical imaging tests is one of the main tasks that radiologists do. For years, it has been a challenge to teach computers to do this kind of cognitive task; the main objective of the field of computer vision is to overcome this challenge. Thanks to technological advances, we are now closer than ever to achieving this goal, and radiologists need to become involved in this effort to guarantee that the patient remains at the center of medical practice.
This article clearly explains the most important theoretical concepts in this area and the main problems or challenges at the present time; moreover, it provides practical information about the development of an artificial intelligence project in a radiology department.
La interpretación de la imagen médica es una de las principales tareas que realiza el radiólogo. Conseguir que los ordenadores sean capaces de realizar este tipo de tareas cognitivas ha sido, durante años, un reto y a la vez un objetivo en el campo de la visión artificial. Gracias a los avances tecnológicos estamos ahora más cerca que nunca de conseguirlo y los radiólogos debemos involucrarnos en ello para garantizar que el paciente siga siendo el centro de la práctica médica.
Este artículo explica de forma clara los conceptos teóricos más importantes de esta área y los principales problemas o retos actuales; además, aporta información práctica en relación con el desarrollo de un proyecto de inteligencia artificial en un servicio de Radiología.
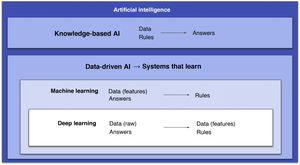
Artificial intelligence (AI) is defined as the capacity for machines to perform intellectual tasks usually performed by humans.1 This term is used as a general concept that encompasses both machine learning (ML) and deep learning (DL). Both concepts belong to a subfield of AI characterised by creating systems that are capable of learning — i.e., generating their own rules using only data (data-driven artificial intelligence).2
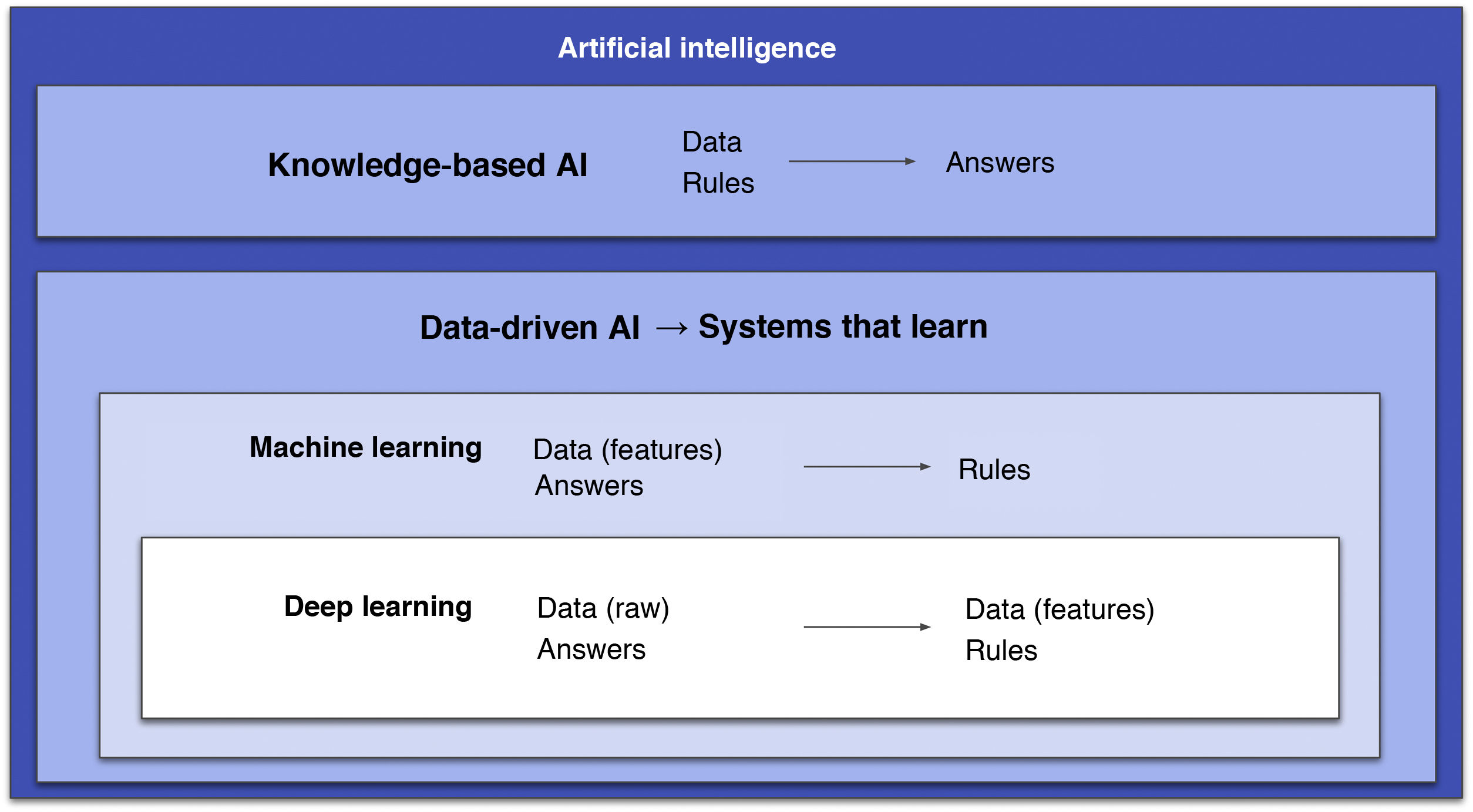
Some authors distinguish between ML and DL based on the fact that the former involves human intervention to train the algorithm through data manipulation (extraction and selection of the most important features), whereas in DL human intervention is minimal as there is no such prior step.2 However, a more correct approach is to consider DL an outgrowth of ML: DL systems are ML systems, but deeper (hence the name). That is, they consist of many more layers, and these extra layers are precisely what grants them the capacity to extract the most relevant features from the data on their own (Fig. 1).2 In this field, a feature is a variable or property of data that can be measured (e.g., pixel value or patient age). The most important features will be the ones that help to solve a given problem. Another important concept is the ML or DL learning system model, as there are three different types (Fig. 2).3
Diagram that encompasses the different types of artificial intelligence (AI). Two main fields are distinguished. One is knowledge-based AI, consisting of systems that require prior programming of rules. The other is data-driven AI, consisting of systems that learn, including machine learning, with algorithms that must be supplied with clean data and, within this, deep learning with systems that, thanks to their larger quantities of layers, are capable of doing this on their own.
AI: artificial intelligence.
AI, and DL in particular, have played a central role in countless articles in recent years, many related to radiology. However, these concepts are not as novel as they are thought to be. In reality, AI arose in the mid-1950s4 and, over the course of history, has sometimes stagnated (during so-called AI winters) and sometimes rebounded. We are currently experiencing an unprecedented rebound, primarily thanks to the development of the technology needed for optimal AI functioning, such as graphical processing units. In the medical sphere, radiology is one of the specialisations that is being revolutionised the most by these new systems.5
Key milestones in the development of artificial intelligenceSince the late 1950s, a series of AI-related successes have had a major media impact.
In 1970-1976, the four-colour theorem, an unsolved mathematical problem, was finally proven with the help of a computer. Thus it became the first example of including computers in solving human problems.6 In that same decade, the term augmented intelligence emerged to describe the use of computers dedicated to enhancing human cognition.7
Later, in 1997, IBM's Deep Blue computer managed to defeat world chess champion Garry Kasparov in a chess tournament. Deep Blue possessed information from thousands of prior games and was capable of analysing all possible situations from subsequent moves.6–8 This was knowledge-based artificial intelligence (Fig. 1), since these systems did not learn anything; rather, they simply applied the rules of the game programmed by humans, with the advantage that computers have of being able to process large amounts of data in little time.8
In 2015, the AlphaGo system, developed by Google DeepMind, became the first system to defeat one of the best players of go, a game that has simple rules but is more complex than chess when it comes to strategy.9 This system was already based on ML techniques implemented through DL neural networks, since the algorithm was capable of inferring or learning on its own the rules of the game based on prior game data.
Closer to the present, in 2017, AlphaZero made its debut. It, unlike AlphaGo, is capable of learning by playing against itself. In other words, it is based on reinforcement learning and is not supplied with data from previous games in advance, thus avoiding any human intervention. With just a few hours of autonomous training, this algorithm was capable of winning go against other programs and prior versions.10
At present, the most successful systems in medical and scientific settings are deep neural networks, generally trained by means of supervised learning.11 These networks are categorised as DL as they learn directly from data with no need for said data to have been pre-selected by humans. The development of these techniques has brought about a paradigm shift in this field, especially in image analysis; hence, these techniques are the focus of this article.
Neural networksNeural networks are predictive models, meaning that given some data in advance they are capable of generating a prediction when presented with new data. Other, better known predictive models are simple linear regression, multiple linear regression and logistic regression. Neural networks yield better results than those models when confronted with more complex problems. In general, predictive models can be divided into classification predictive models and regression models. Classification models are based on finding a discrete prediction for the input variable, such as predicting the presence or absence of a specific disease based on an image. Regression models, on the other hand, are used to find continuous predictions for the input variable, such as predicting D-dimer levels based on input variables (namely, age, presence of cancer, etc.). The foundations of neural networks are explained below, starting with artificial neurons and ending with convolutional neural networks, which constitute the type of neural network that has demonstrated the most success in artificial vision.
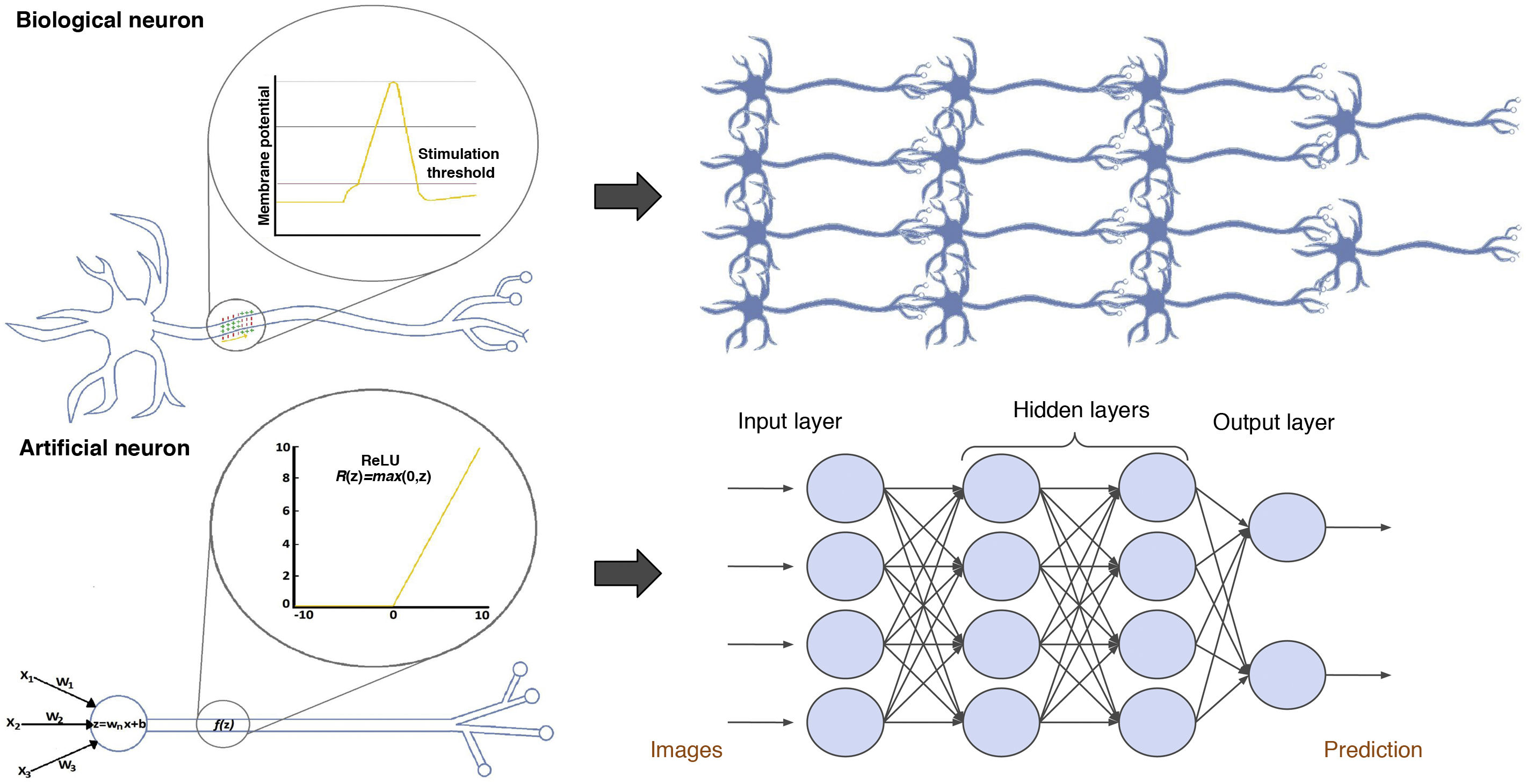
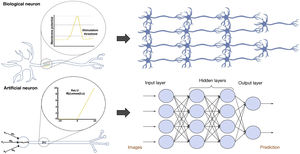
Artificial neuronsArtificial neural networks consist of multiple interconnected artificial neurons, also called simple perceptrons, which can be compared to biological neurons (Fig. 3). An artificial neuron or simple perceptron consists of several input routes, which resemble dendrites of biological neurons and transmit information to the soma. The soma of an artificial neuron is a function that integrates all the information from the inputs and, after applying an activation function, generates an output.3
Comparison between biological and artificial neurons and between biological and artificial neural networks. Artificial neural networks are divided into three main parts: the input layer is a layer of perceptrons specialised in receiving information; the hidden layers are layers capable of extracting features from data and transforming them in pursuit of the best representation of the problem to be solved; and the output layer is a layer prepared to offer the output information, such as the class to which the input image belongs according to the network's prediction in classification problems.
An activation function might resemble the biological process of membrane depolarisation, which, instead of following a linear function, answers to the all-or-none law. Biological neurons receive many impulses that do not manage to activate or depolarise them, until one arrives with sufficient potential to depolarise it and thus generate an output or action potential that travels through the axon, which will transmit the impulse to the contiguous neurons.12 Artificial neurons also receive a number of stimuli, and if any of them manages to activate the activation function, this will give rise to an output. The mathematical reasoning that explains why these functions are indispensable is that they are responsible for introducing non-linearity into the neuron, which makes it possible to approach much more complex functions and thus solve, for example, classification problems with clusters that cannot be separated by lines.3
Classic artificial neural networksBiological neurons are arranged in layers to form biological neural networks. Artificial neural networks do the same by forming artificial networks; therefore, the arrangement of perceptrons in layers and the concatenation of successive layers are what form a neural network (Fig. 3).13
The architecture of deep neural networks may resemble the biological model of the primary visual cortex proposed by DH Hubel and T. Wiesel, both Nobel prize winners, in 1959. According to this biological model, the primary visual cortex consists of two types of cells: simple cells and complex cells. Simple cells, also called edge detectors, respond positively when they detect the edge of an object in a particular orientation, whereas complex cells leverage the contributions of edge detectors to find all an object's edges. The hierarchical arrangement of these cells in layers means that these objects are recognised sequentially, starting with their simplest features and ending with their most complex ones. Neural networks maintain a similar architecture: the first layer of the network is in charge of extracting the object's coarse-grained features, such as edges and contrasting colours. Information then passes through successive layers which proceed to extract more fine-grained details.14
The learning or training processBefore beginning network training, variables called hyperparameters must be selected. Hyperparameters are variables that determine the structure of the network and how it is trained; hence, they are defined before starting training and are adjusted over time based on the results thereof. The type of activation function used and the number of hidden layers in the algorithm are examples of hyperparameters.2
The learning or training process for a neural network consists of adjusting certain parameters called weights. Weights are understood as intensities of connections existing between artificial neurons. In a biological neuron, weights could resemble intensities of synapses between neurons. Thus, adjusting synapses achieves optimal end results.15
Each time an image enters the network, all the neurons in all the layers in the network are sequentially activated such that they generate weights for each neural connection in what is called forward propagation. Finally, on arrival at the last layer, through this process, a prediction is generated for that image. However, given that the network is still being trained, how can that prediction be known to be right or wrong? How can the network be made to continue improving with each training image?
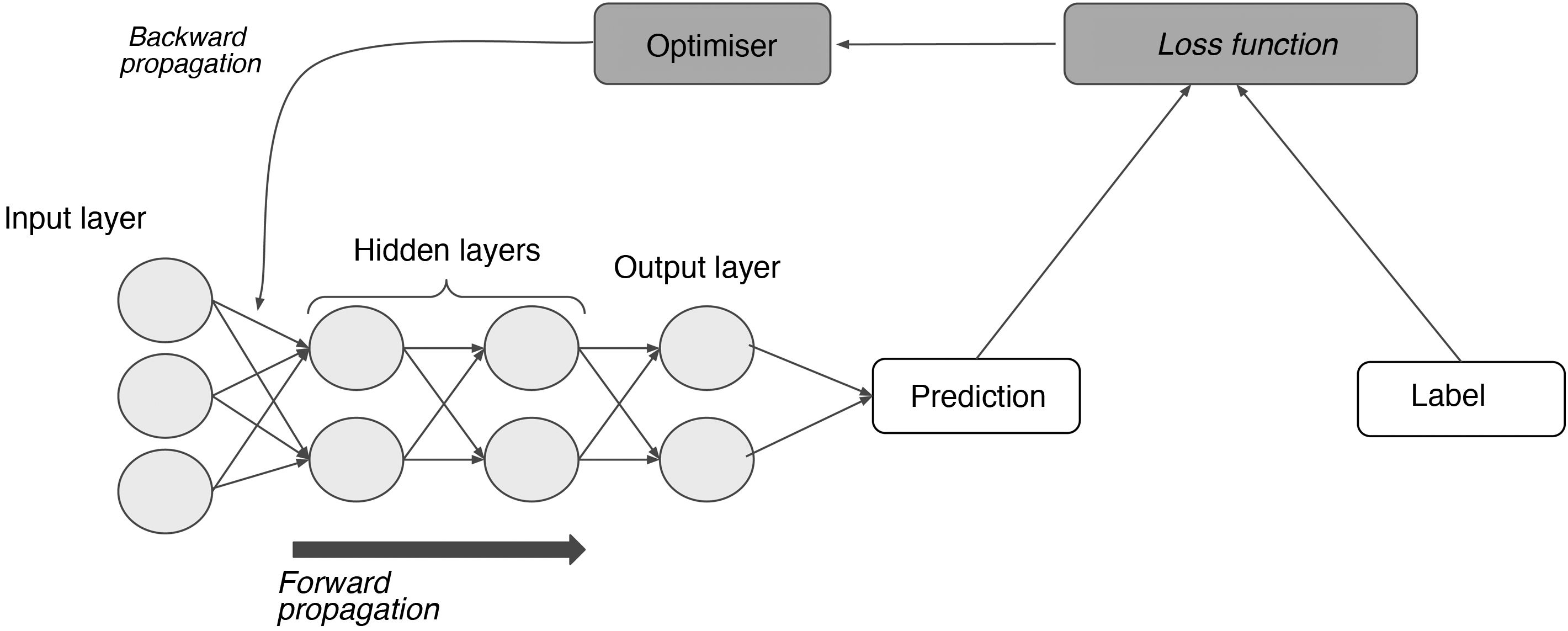
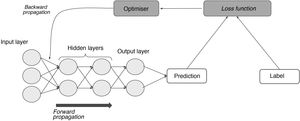
In a supervised learning model, the neural network starts out using some random weights and learns when those weights are adjusted by comparing the network's results for an example to the labelled results.11 This requires, on the one hand, labels and, on the other hand, a function that measures the error generated (loss function); an optimisation algorithm that calculates magnitude by which and direction in which weights must be modified with the goal of minimising that error (gradient descent); and, finally, another function capable of feeding that adjustment back through the network and modifying each neuron's weights based on the extent to which that neuron is responsible for the end result (backward propagation) (Fig. 4).15
The process of supervised learning (training). Through the loss function, the difference between the network's prediction and the label for each input is measured; next, through backward propagation of this error and of the optimisation algorithm, the weights of the different neurons are adjusted until they correspond to a minimum in the loss function.
This weight adjustment process is called network learning and occurs primarily in the hidden layers. Throughout the network, through weight adjustment, the hidden layers form increasingly complex representations of data, but become increasingly adjusted to the problem (Fig. 5).2 Thus, the foundation of both ML and DL consists of successively transforming data until the best representation that enables the problem to be solved is found. The term deep refers not to a deeper understanding of data, but to learning successive, increasingly meaningful representations, and the number of these layers is what is known as the depth of the model.3
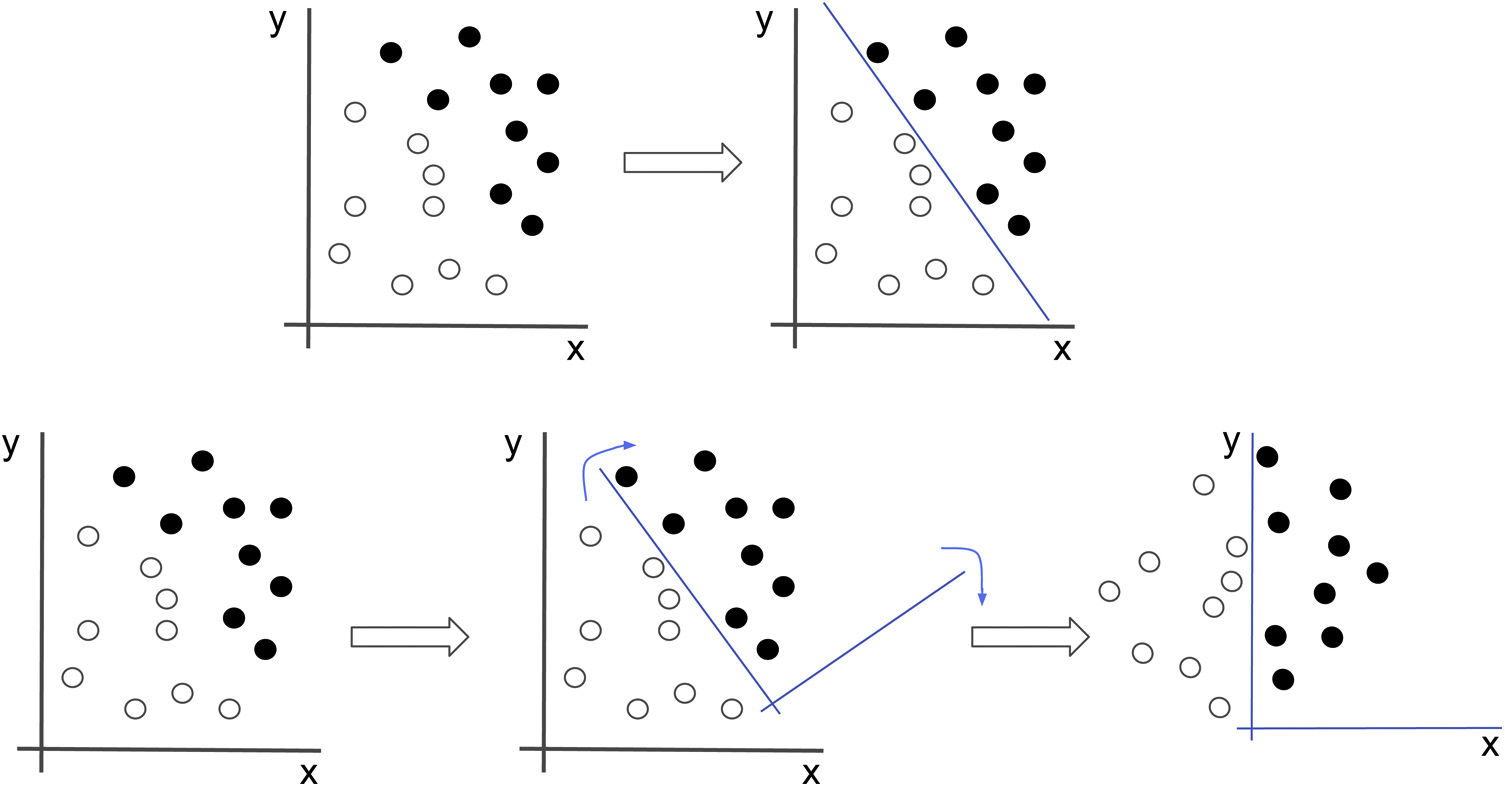
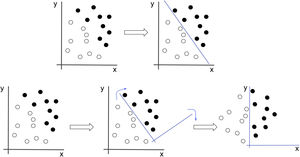
Neural networks seek the best representation of the data that allows them to solve the problem. In this example, if we were to attempt to classify the black and white dots, we would have to draw a line which would correspond to a non-intuitive equation. If, however, we apply a transformation to the data that causes the image to rotate, the problem suddenly becomes much simpler (x=0).
To train a neural network, there must be at least two subsets of data: a training set, with which the model will adjust its weights according to a minimum in the loss function; and a validation set with which to evaluate the yield thereof. Iterations (called epochs) on these datasets and the model will yield better and better results which will be observed in the evaluation of the model's performance with the validation set in each iteration. If the model's performance is poor, the expert may make changes to the hyperparameters.
Finally, once training is finished — i.e., once both the weights and the hyperparameters have been adjusted — the model is tested with new data (a test set) to evaluate its true performance. In this process, the model is exposed to new data, such as unlabelled images, and a prediction — for example, the class to which that image belongs in a classification problem — is obtained. Test data must never be used to modify the model's weights or hyperparameters.
Convolutional neural networksWith the expanding use of classic neural networks, problems began to arise that drove the development of more complex forms of neural networks. In the case of imaging and object recognition, the main problem was that, generally, the same object could have different shapes and be in different positions, and this would reduce the networks' performance. Thus convolutional neural networks (CNNs) emerged. These are the ones that are most commonly used for medical imaging.15
The above-mentioned classic neural networks consist of completely connected layers. This means that all the neurons in a layer are connected with those of the next layer, and therefore the image is interpreted in its entirety, taking as an input the value of all the pixels and performing operations that include all the image's information. If, for example, the network has a goal of learning to identify cars, and a car appears in the upper left-hand corner of one image and the lower right-hand corner of another, the network must learn different weights for each of those images, as the different location of the same object leads to its interpretation as different objects, each with its own specific weights and representations. Consequently, these networks do not perform well and are inefficient at tasks such as image interpretation and object identification. CNNs, by contrast, have matrices called filters that are capable of analysing the composition of an image and granting the network the capacity to identify a car regardless of its location. This renders them much more efficient at image interpretation than classic neural networks.15
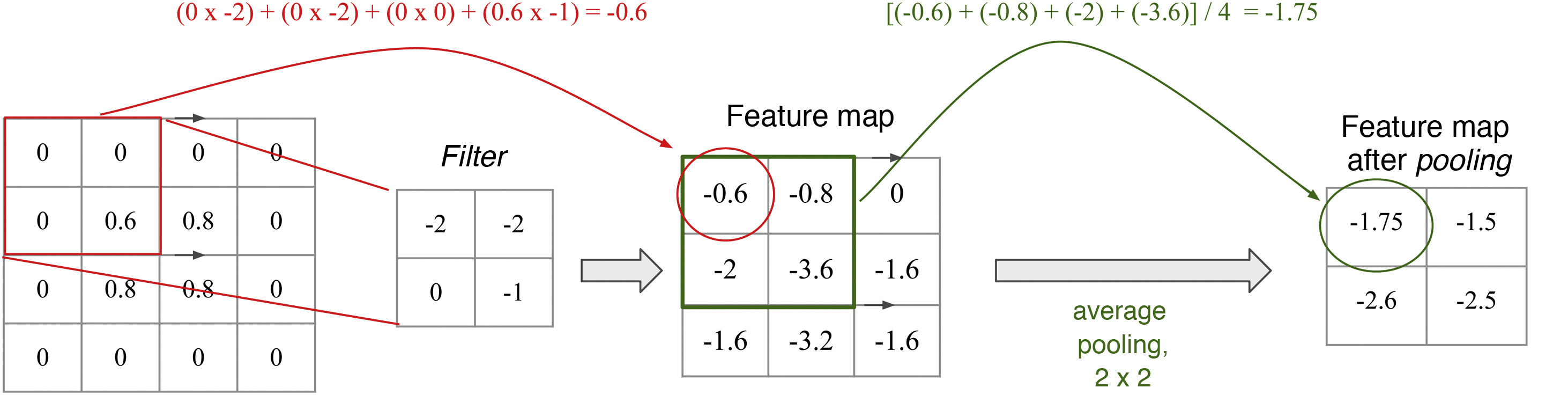
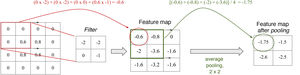
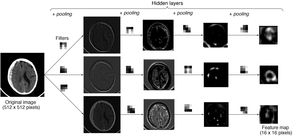
Each convolutional layer of a CNN can consist of multiple filters. These filters are numerical matrices that scan the image and perform convolutional operations on groups of pixels to produce feature maps. Each filter represents a feature (Fig. 6). Thus, layer after layer, increasingly complex features are extracted and increasingly coarse-grained representations of the input data are formed. It is normal for convolutional layers to be followed by pooling layers, which reduce map dimensionality and thus computational cost (Fig. 7). The representations of the last layer of the convolutional part are transformed into a final vector through one or more completely connected layers and, finally, into a prediction. This second part of the network is commonly called the classifier.15
Filters and pooling: A filter is a matrix that scans the image and performs a multiplication operation element by element and then an addition operation to yield a value (convolution). High values are obtained when the filter is applied to a feature similar to the filter; low values are obtained when it is applied to a feature that is different from it. In a filter that detects vertical edges, high values mean that a vertical edge has been detected. Is this not reminiscent of the cells of the primary visual cortex? The pooling layer, average with a size of 2 × 2 in this example, determines the mean of the values in the range, thus reducing the dimensionality of the feature map.
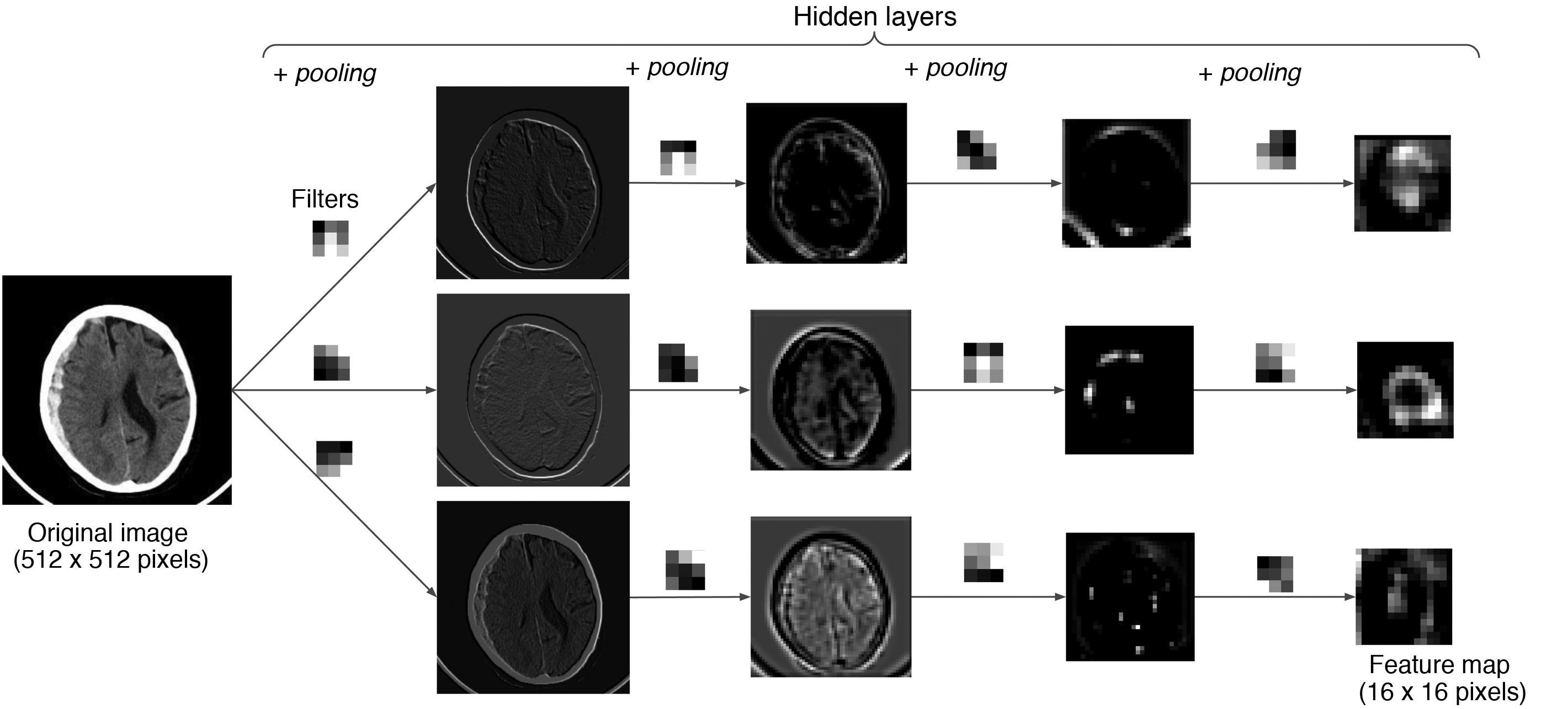
Example with an image: an example is shown in which an image from a CT scan of the brain passes through successive layers that contain three 3 × 3 filters and the pooling function, yielding different feature maps with lower and lower spatial resolution. The greater the depth in the network, the more coarse-grained and smaller the maps, with forward propagation of only the most important information.
In conclusion, CNNs, thanks to filters, learn local patterns, and therefore are capable of recognising these patterns regardless of whether repositioning is performed, while classic neural networks learn overall patterns and are not capable of abstraction from the location, orientation or shape of the object in the image. Thus CNNs have proven most suitable for working with medical imaging, as they are capable of performing such complex tasks as image classification.2
Main problems with neural networks and some solutionsNotable among the obstacles that may be encountered while training a neural network that can be detected using the validation set are overadjustment and underadjustment. Overadjustment occurs when the model is so specialised in training data it is incapable of generalising and it does not yield good results when presented with new data. Underadjustment refers to a model that, due to insufficient specialisation or excessive simplicity, is incapable of yielding good results even with training data. In both cases, the model will not have found the important features that would enable it to solve the problem in general or with new data. That is, in both cases, the model will not have learned to generalise. Ultimately, neural networks must learn transformations, not specific examples.3
Overadjustment is directly tied to one of the most important obstacles encountered in developing medical imaging-related systems: a shortage of labelled data. One of the reasons for this problem is that creating extensive databases of duly labelled images requires a great deal of time and effort on the part of experts — in our case, radiologists. In addition, as an imaging diagnosis is not always consistent with a histological diagnosis, the question of which test should be considered diagnostic of the disease to be studied so that the label may be created must be weighed carefully. Imaging is considered diagnostic in some conditions, such as fractures. However, most diseases require other evidence to make a definitive diagnosis, be it results of histological tests or clinical/laboratory findings. For example, in most cancer cases, histological results are needed for diagnosis.16 This problem of a dearth of labelled images is often exacerbated by the complicated ethical and legal framework for transmitting medical data.17
Nevertheless, there are several projects under way with the objective of creating extensive databases with labelled medical images, such as the Cancer Imaging Archive.18 There are also companies like Savana19 that offer AI solutions to leverage medical data in free-text format. In addition, strategies are being put forward such as interactive reports, in which the radiologist can create hyperlinks to other texts or labels in the report itself;16 structured reports; and even international collaboration projects involving many radiologists, such as the preparation of the dataset for the RSNA 2019 Brain CT Hemorrhage Challenge;20 platforms like OpenNeuro,21 which provides access to databases of both brain imaging and electroencephalograms; the European Network for Brain Imaging of Tumours (ENBIT);22 and consortia like ENIGMA,23 which brings together researchers in genomics and brain imaging.
Another one of the solutions demonstrating the greatest success, primarily in medical settings, is transfer learning. This technique consists of being able to transfer to a given model, from a pre-trained network, both the architecture and the weights of the first layers. The weights of the first layers have been chosen for transfer due to the hierarchical learning of neural networks, discussed above, according to which the first layers are responsible for extracting simpler features, i.e., features that are less specific to the problem to be solved and presumed to be common to both sets of images. In this regard, one can either train solely the last part of the network, the classifier, and keep the convolutional part frozen, or also train a variable number of layers of the convolutional part, which is called unfreezing layers. In any case, this new model begins its learning process with an advantage, as it must adjust weights from a favourable position instead of starting with random values. This is why these models can yield better results with fewer data than completely new models.24
Finally, one of the most significant and, at the same time, most difficult-to-solve problems with these systems is their limited transparency. Although the mathematical process by which the algorithms are constructed can be explained, how they reach their conclusions is not clearly understood. Even now, CNNs are considered black boxes,25 and research is needed to improve their explainability. One very commonly used solution is gradient-weighted class activation mapping (Grad-CAM).26 Grad-CAM systems use a gradient to locate the areas of the image on which the algorithm focuses to make a final decision. At the same time, this limited explainability and transparency hamper the development of an ethical and legal framework for regulating the implementation of these systems in regular medical practice.27
Development of artificial intelligence systems in radiology departmentsIn a diagnostic imaging department, AI systems can be applied in multiple areas, such as performing tasks related to patient scheduling;28 selecting optimal imaging protocols and radiation doses;29 placing patients in equipment;30 performing image post-processing (reconstructions, best image quality, etc.);31 and, of course, as explained above, interpreting images.32 In the latter regard, efforts are under way to develop not only AI systems that make diagnoses, but also systems capable of segmenting organs and detecting lesions, as well as monitoring them.33 Going a bit further, research is being conducted on systems that predict, for example, estimated survival or disease severity based on lesion type or other patient clinical/laboratory data.32–34 Adding patient clinical data to imaging data can bring about substantial improvements in the results of these AI models, which has led to the creation of hybrid networks — networks that combine DL and ML methods.34
When the development of a medical imaging interpretation-related AI system is proposed, the approval of the hospital's independent ethics committee must first be obtained. Generally, for retrospective studies in which obtaining informed consent is not feasible and the risks of a medical data breach are minimal, acquiring the patient's informed consent is usually not necessary.35
Next, patients are selected for inclusion in the study and their imaging is collected. This is one of the most important steps in the development of these systems, which, as mentioned above, largely depend on data quantity and quality (data-driven systems). To date, as labelling of radiological imaging is not a widespread practice and as radiology information systems are not prepared for this type of search, obtaining images corresponding to a specific disease is no easy task.35 Here, natural language processing offers techniques capable of collecting structured data from radiology reports, with very promising results.36 Also very important is image de-identification which, in the case of images in DICOM format, can be complex. Once data are collected, they can be preprocessed based on the type of neural network to be trained.16
Once the database has been prepared, it is divided into the three above-mentioned subsets: the training set, the validation set and the test set, accounting for approximately 80%, 10% and 10% of the total, respectively.16
We offer a GitHub repository featuring an example of a neuron that solves a regression problem and an example of a neural network that solves a classification problem, at the following link: https://github.com/deepMedicalImaging/RedNeuronalArtificial.git.
Authorship- 1
Responsible for the integrity of the study: AP, PM, PS, LL and DR.
- 2
Study concept: AP, PM, PS, LL and DR.
- 3
Study design: AP.
- 4
Data collection: not applicable.
- 5
Data analysis and interpretation: not applicable.
- 6
Statistical processing: not applicable.
- 7
Literature search: AP.
- 8
Drafting of the article: AP.
- 9
Critical review of the manuscript with intellectually significant contributions: AP, PM, PS, LL and DR.
- 10
Approval of the final version: AP, PM, PS, LL and DR.
The authors declare that they have no conflicts of interest.
Please cite this article as: Pérez del Barrio A, Menéndez Fernández-Miranda P, Sanz Bellón P, Lloret Iglesias L, Rodríguez González D. Inteligencia artificial en Radiología: introducción a los conceptos más importantes. Radiología. 2022;64:228–236.