Reverse transcriptase - polymerase chain reaction (RT-PCR) is the standard technique for SARS-CoV-2 diagnosis. The World Health Organization recommends the Charité-Berlin protocol for COVID-19 diagnosis, which requires triple PCR, limiting the process capability of laboratories and delaying the results. In order to reduce these limitations, a duplex PCR is validated for the detection of the E and ribonuclease P genes.
MethodsWe compared the limit of detection, sensitivity and specificity of the duplex PCR technique (E gene and Rnasa P) against the monoplex standard (E gene) in RNA samples from a SARS-CoV-2 isolate and 88 clinical specimens with previously known results. The repeatability and reproducibility of the threshold cycle values (Ct) were determined in two independent laboratories of the Faculty of Medicine of the Universidad de Antioquia, using different reagents and real time instruments.
ResultsThere were no significant differences in the Ct results between both techniques (P = .84). Using the monoplex PCR of E gene as a reference, the interrater reliability analysis showed similarity between the two techniques, with a kappa coefficient of 0.89, the sensitivity and the specificity of duplex PCR were 90% and 87%, respectively.
ConclusionsDuplex PCR does not affect the sensitivity and specificity reported by the Charité, Berlin protocol, being a useful tool for SARS-CoV-2 screening in clinical samples.
El estándar de diagnóstico para SARS-CoV-2 es la reacción en cadena de la polimerasa (PCR). La Organización Mundial de la Salud recomendó el protocolo de Charité-Berlín para el diagnóstico de COVID-19; esta metodología implica tres PCR, limitando la capacidad de procesamiento y retrasando los resultados. Con el fin de reducir estas limitaciones, se validó una PCR dúplex para la detección del gen E y ribonucleasa P.
MétodosSe comparó el límite de detección, sensibilidad y especificidad de la técnica de PCR dúplex (gen E más RNasa P), comparada contra el estándar monoplex (gen E) en muestras de ARN de un aislado de SARS-CoV-2 y de 88 especímenes clínicos con resultados previamente conocidos. Se determinó la repetibilidad y reproducibilidad de los valores de ciclos umbrales (Ct, cycle threshold), en dos laboratorios independientes de la Facultad de Medicina de la Universidad de Antioquia, usando reactivos y equipos diferentes.
ResultadosNo hay diferencias significativas (P = ,84) en los resultados de Ct entre ambas estrategias. Al utilizar como referencia el gen E amplificado en Monoplex, el análisis de concordancia demostró fuerte similitud entre las dos estrategias, con un coeficiente kappa de Cohens de 0.89, una sensibilidad del 90%, y una especificidad del 87%.
ConclusiónLa PCR dúplex no afecta la sensibilidad y especificidad informadas por el protocolo Charité, Berlín, siendo una herramienta útil para el cribado de SARS-CoV-2 en muestras clínicas.
At the end of December 2019, in the city of Wuhan in the province of Hubei, China, an alert was generated following the diagnosis in different hospitals of patients with symptoms of pneumonia of unknown origin after these cases were epidemiologically related to frequenting and consuming food from a wholesale seafood market1. By late 2019 and early 2020, the Chinese Centre for Disease Control and Prevention (China CDC) identified the cause of the disease as a new coronavirus, which was named by the International Committee on Taxonomy of Viruses in February 2020 as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), and as of 11 February 2020 the disease was called coronavirus disease 2019 (COVID-19)2.
Currently (up until 10 November 2020), there are nearly 40,000 SARS-CoV-2 sequences published in the GenBank database, including the complete genomic sequence of the new agent. This has enabled a number of different diagnostic protocols to be developed, including real-time polymerase chain reaction (PCR) with probes and primers widely used for the detection of SARS-CoV-2.
On 17 January 2020, the World Health Organization (WHO) published an update of the approved protocols for performing laboratory tests3, and on 1 February of the same year, the Pan American Health Organization (PAHO) adopted the protocol developed by the Charité Hospital, Berlin, Germany, as the diagnostic method for the detection of SARS-CoV-24. This guideline was subsequently adopted by the Instituto Nacional de Salud de Colombia [National Institute of Health of Colombia].
The Charité-Berlin protocol diagnoses SARS-CoV-2 by amplifying and detecting a region of the virus envelope (E gene) shared by different betacoronaviruses of the Sarbecovirus subgenus as a screening test, and in samples identified as positive, performing a confirmatory PCR to detect a specific region of SARS-CoV-2 located in the RNA-dependent RNA polymerase (RdRp) gene5. In addition, following good laboratory practice, the amplification of a human control gene is included to identify the viability of the sample and PCR inhibitors and to evaluate the efficiency of RNA extraction; the gene most used for this purpose is the ribonuclease P (RNase P) gene.
According to the above protocol, three independent PCR have to be performed to diagnose SARS-CoV-2. However, this delays the diagnosis, as it takes longer to deliver the results and means greater use of reagents, which increases the cost of the test.
The high demand for reagents worldwide and the fact that it takes over 30 days to import these items to Colombia, compounded by the increase in new cases in recent weeks, mean that we have to make rational use of the tests and develop methods to optimise available resources.
The implementation of a duplex PCR between the E and RNase P genes could reduce the time to obtain test results and increase laboratories' processing capacity.
The objective of this study was to validate a duplex PCR technique for the diagnosis of SARS-CoV-2 based on the detection of the E gene and RNase P.
MethodsSARS-CoV-2 cell culture and clinical specimens. RNA was extracted from the SARS-CoV-2 strain isolated and cultured by the Immunovirology group at the University of Antioquia6, and this served as a positive control to standardise and evaluate the performance of the duplex PCR.
We selected 88 RNA samples from clinical specimens taken from samples of aspirate and nasopharyngeal swabs from patients with suspected COVID-19 who had a previous result for SARS-CoV-2. Duplex PCR was performed for the samples at the Laboratorio Integrado de Medicina Especializada (LIME) [Integrated Laboratory of Specialised Medicine] and the Immunovirology Group (IMV), both in the Faculty of Medicine at the University of Antioquia, in order to evaluate the repeatability and reproducibility of the technique using different reagents and equipment.
RNA extraction. For the virus isolate, extraction was performed from culture supernatants using the SaMag-12 automated extraction system and the SaMag™ Viral Nucleic Acid Extraction Kit (Sacace Biotechnologies, Italy), following the manufacturer's instructions, with an elution volume of 30 μ.
RNA extraction from clinical specimens was performed from nasopharyngeal aspirate or nasopharyngeal swab samples from patients with suspected COVID-19 using two automated systems with Magmax magnet technology for RNA extraction (Thermo Fisher Scientific Inc, USA) and SaMag™ (Sacace Biotechnologies, Italy) at LIME and IMV respectively. In addition, manual extractions were performed with the Quick-RNA Viral Kit (Zymo Research, USA), in accordance with the manufacturer's instructions. The elution volume for the automated kits was 30−50 μL and for the manual kit, 15 μL. It should be clarified that extraction was performed by different methods with the aim of expanding the laboratory's diagnostic capacity, and depending on the availability of reagents in Colombia.
The RNA was stored at −80 °C until use and the same product extracted from each sample was used for all RT-PCR reactions to be analysed in this article.
Primers and probes. The primers and probes used were those for the Sarbeco E and RdRp genes, published by Corman et al. in the Charité-Berlin protocol5. The RNase P gene was used as an internal reaction control and was amplified using two pairs of priming oligonucleotides: 68 samples were amplified with the oligonucleotides and probe published by the CDC for SARS-CoV-2 detection7 and 20 samples were amplified using the CDC probe, but with a pair of primers designed at the binding site of exons 1 and 2 of the RNase P gene (RNase-P-Fw: 3′-ATGGCGGTGTTTGCAGATTTG-5′ and RNase-P-Rv: 3′-CAACTGAATAGCCAAGGTGAGC-5′) to ensure amplification of the extracted RNA, excluding genomic DNA.
The E and RdRp probes were labelled with the FAM fluorophore, and VIC or HEX were used for RNase P depending on the compatibility of the real-time PCR equipment used.
qRT-PCR assays for the detection of SARS-CoV-2. The reverse transcription and subsequent amplification of the SARS-CoV-2 viral genome in real time (qRT-PCR) were performed using the enzyme SuperScript™ III One-Step RT-PCR System (Invitrogen™, USA) and the Luna® Universal One-Step RT-qPCR Kit (New England Biolabs, USA). The E gene was amplified individually (monoplex) and in duplex with RNase P; the RdRp gene was only amplified in monoplex.
The PCR amplifications for the virus genes RdRp and E (both in monoplex and duplex) were performed following the recommendations for concentrations of oligonucleotides (primers and probes) published in the Charité-Berlin protocol5; RNase P was amplified in the PCR duplex with an oligo concentration of 0.15 μM and probe at 0.2 μM.
The thermal cycling conditions for the SuperScript™ III One-Step RT-PCR System enzyme were those published in the Charité-Berlin protocol5, and when the Luna® Universal One-Step RT-qPCR Kit enzyme was used, the reverse transcription was modified to 55 °C for 18 min and alignment/extension to 60 °C for 30 s, following the manufacturer's recommendations.
The samples were considered positive when the fluorescence exceeded the detection threshold by a Ct less than 38 and gradually increased over the cycles, generating a sigmoidal amplification curve. Samples with a Ct greater than 38 were considered negative.
Comparison of the limit of detection between monoplex PCR of E gene, monoplex of RdRp and duplex (E gene and RNase P)RNA extracted from the SARS-CoV-2 strain isolated by the Immunovirology Group was used for the comparison of performance between the duplex assay and the monoplex assay. Serial decimal dilutions of viral RNA were performed, using RNA extracted from a sample negative for SARS-CoV-2 as diluent, ending with a 1 × 10−8 dilution. Five replicates of the amplifications of the E genes and duplex E plus RNase P genes were performed: three at LIME and two by the Immunovirology group. The PCR of the RdRp gene was performed in triplicate at LIME.
The reproducibility of the results between laboratories was evaluated by comparing two RNA extractions from the viral culture independently diluted from 1 × 10−1 to 1 × 10−8; duplicate PCR of the E gene and duplex E plus RNase P genes were performed in each dilution series.
Finally, to evaluate the performance of duplex PCR in clinical samples, 88 RNA samples were selected from patients with suspected COVID-19 (39 positive and 49 negative for SARS-CoV-2) and analysed in the 7500 Fast (Applied Biosystems, USA) and CFX-96 (Biorad) thermal cyclers, used by LIME and IMV, respectively.
Statistical calculations. The threshold cycles (Ct) of samples identified as positive for SARS-CoV-2 from the monoplex and duplex amplifications were compared using the paired t-test in the Statistics Kingdom software (https://www.statskingdom.com). A P value <.05 was considered significant.
The SARS-CoV-2 results from the clinical samples obtained in monoplex and duplex were compared using Cohen's kappa coefficient. A kappa value between 0.81 and 1 is considered as “almost perfect” agreement8.
Sensitivity and specificity values were obtained using the Medcalc software (https://www.medcalc.org/).
ResultsComparison of performance between monoplex PCR and duplex PCRThree independent PCR were performed: E gene and RdRp in monoplex, and E gene plus RNase P in duplex, of the serial dilutions of the viral RNA from cell culture using RNA extracted from a SARS-CoV-2 negative sample as diluent.
The concentration of the viral culture at the time of extraction was 4.2 × 106 PFU/mL which, according to the quantification of other SARS-CoV-29 isolates, corresponds to an approximate RNA concentration of 4 × 109 genome equivalents/mL, and it was diluted serially to 1 × 10−9; subsequently, the three PCR were performed simultaneously, with five replicates of each dilution for the E gene and the duplex E gene plus RNase P (three replicates performed by the LIME laboratory and two replicates by Immunovirology), while the RdRp gene was amplified in triplicate at LIME.
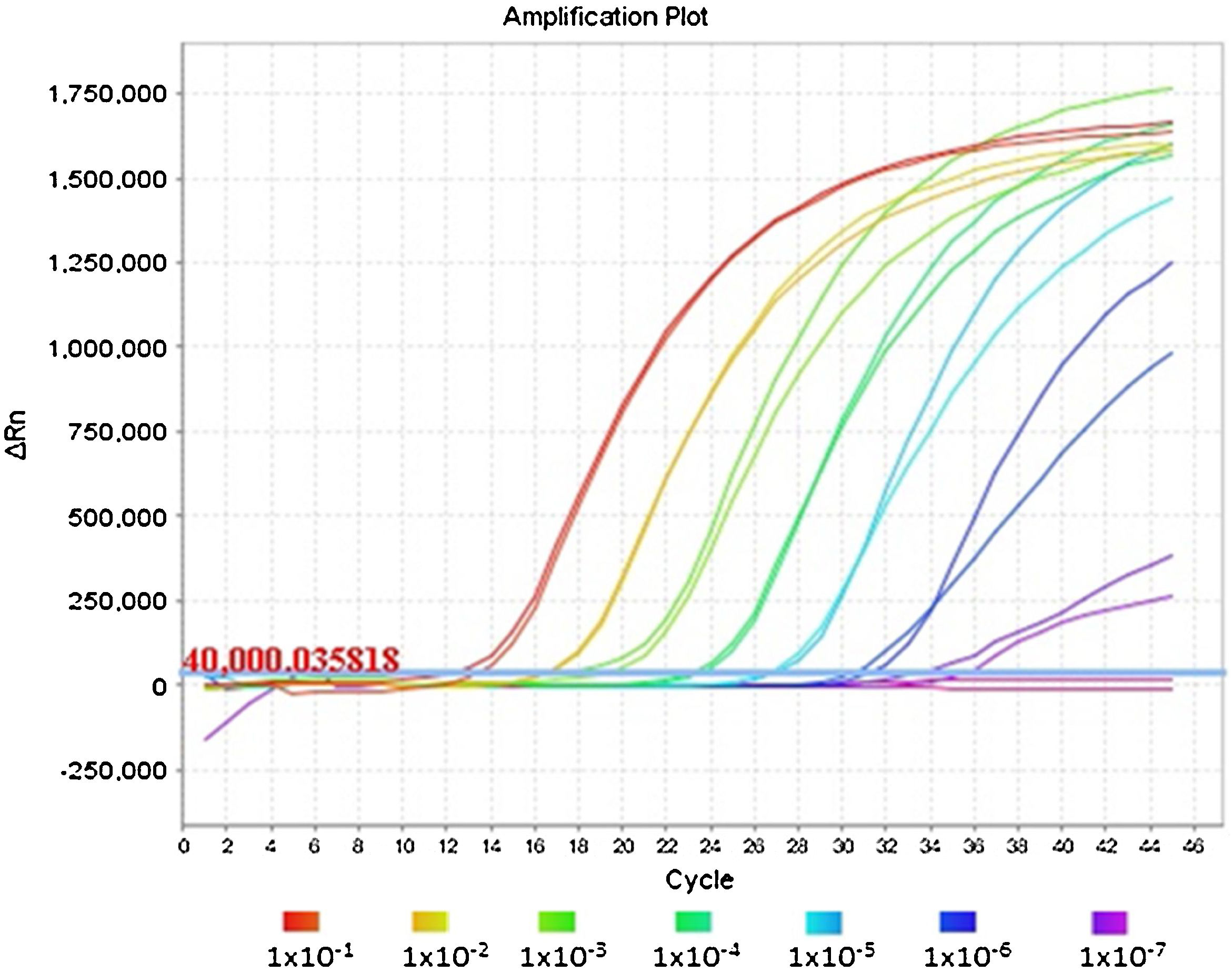
There were no significant differences in the Ct results (P value = .7662) between the monoplex of the E gene and its combination with RNase P (Table 1). In fact, the averages and standard deviations of the Ct were similar between groups at all dilutions and detection was identified in all five replicates of the 1 × 10−7 dilution, corresponding to 0.42 genome equivalents/μl. This demonstrates the repeatability and reproducibility of the results.
Ct values of serial dilutions of viral RNA from cell culture diluted in human RNA negative for SARS-CoV-2.
| Technique | Monoplex | Duplex | Monoplex | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dilution | E gene | E gene | RNase P | RdRp gene | ||||||||
| Ct | Average | SD | Ct | Average | SD | Ct | Average | SD | Ct | Average | SD | |
| 1 × 10−1 | 13.2 | 12.9 | 0.5 | 12.6 | 12.8 | 0.4 | 0 | 28.4 | 0.1 | 14.3 | 14.7 | 0.3 |
| 13.6 | 13.4 | 0 | 14.9 | |||||||||
| 12.9 | 12.7 | 0 | 14.9 | |||||||||
| 12.4 | 12.4 | 28.3 | ||||||||||
| 12.3 | 12.7 | 28.4 | ||||||||||
| 1 × 10−2 | 15.0 | 16.0 | 0.8 | 16.7 | 15.7 | 0.8 | 33 | 29.4 | 2.2 | 18.9 | 18.8 | 0.1 |
| 16.9 | 14.5 | 28.8 | 18.8 | |||||||||
| 16.9 | 15.9 | 30.1 | 18.7 | |||||||||
| 15.6 | 15.8 | 27.9 | ||||||||||
| 15.7 | 15.7 | 27.5 | ||||||||||
| 1 × 10−3 | 16.9 | 17.9 | 1.0 | 19.6 | 19.4 | 0.6 | 28.6 | 27.9 | 0.7 | 22.1 | 22.2 | 0.2 |
| 16.9 | 20.2 | 28.1 | 22.4 | |||||||||
| 17.9 | 18.7 | 28.5 | 22.2 | |||||||||
| 19.0 | 19.2 | 27.1 | ||||||||||
| 18.9 | 19.1 | 27.3 | ||||||||||
| 1 × 10−4 | 24.2 | 22.6 | 1.4 | 23.4 | 23.2 | 0.8 | 27.4 | 27.6 | 0.2 | 25.8 | 25.9 | 0.1 |
| 23.7 | 23.8 | 27.8 | 25.9 | |||||||||
| 20.6 | 24.0 | 27.9 | 25.9 | |||||||||
| 22.3 | 22.5 | 27.6 | ||||||||||
| 22.2 | 22.3 | 27.5 | ||||||||||
| 1 × 10−5 | 27.0 | 26.0 | 1.1 | 26.8 | 26.6 | 0.8 | 27.5 | 28.0 | 0.3 | 29.5 | 29.5 | 0.2 |
| 27.2 | 27.4 | 27.9 | 29.7 | |||||||||
| 24.8 | 27.2 | 28.1 | 29.4 | |||||||||
| 25.3 | 26.0 | 28.2 | ||||||||||
| 25.4 | 25.5 | 28.0 | ||||||||||
| 1 × 10−6 | 30.7 | 30.3 | 1.0 | 30.6 | 29.9 | 1.1 | 27.7 | 27.9 | 0.4 | 33.71 | 33.4 | 1.1 |
| 31.5 | 31.3 | 27.3 | 34.24 | |||||||||
| 30.8 | 29.7 | 28.2 | 32.13 | |||||||||
| 29.1 | 29.8 | 28.1 | ||||||||||
| 29.4 | 28.3 | 28.1 | ||||||||||
| 1 × 10−7 | 31.8 | 33.2 | 1.6 | 33.7 | 33.9 | 3.2 | 26.7 | 27.6 | 0.7 | 40.9 | 39.4 | 2.1 |
| 35.8 | 31.9 | 26.9 | 37.9 | |||||||||
| 32.2 | 38.3 | 28 | 0 | |||||||||
| 32.3 | 35.6 | 28.1 | ||||||||||
| 33.6 | 30.3 | 28.1 | ||||||||||
| 1 × 10−8 | 34.9 | 35.1 | 1.1 | 0 | 38.24 | 2.1 | 27.2 | 27.6 | 0.4 | 0 | 0 | 0 |
| 0.0 | 38.5 | 27.6 | 0 | |||||||||
| 36.3 | 0 | 27.5 | 0 | |||||||||
| 34.2 | 36.1 | 0 | ||||||||||
| 0 | 40.2 | 28.2 | ||||||||||
| 1 × 10−9 | 0 | 0 | 0 | 0 | 0 | 0 | 27.5 | 27.8 | 0.4 | 0 | 0 | 0 |
| 0 | 0 | 27.3 | 0 | |||||||||
| 0 | 0 | 28.0 | 0 | |||||||||
| 0 | 0 | 0 | ||||||||||
| 0 | 0 | 28.2 | ||||||||||
The 1 × 10−8 dilution was amplified in three of the five replicas, where we were able to see the stochastic effects produced by the low viral load of the dilutions and the amplification below the limit of detection of 5.2 copies reported by the Charité-Berlin protocol.5 We should mention that the RdRp gene has lower sensitivity (1 × 10−7 dilution) compared to the E gene (1 × 10−8 dilution), so a specific gene with higher sensitivity should be used for weak positive samples (Table 1).
Fig. 1 shows an optimal amplification of the E gene in the duplex PCR, achieving a sensitivity indistinguishable from that obtained with the monoplex PCR of the E gene when the Ct values are compared. The analysis of the amplification curves of the different dilutions evaluated demonstrates the sigmoidal characteristic in the presence of high concentrations of viral RNA. However, when RNase P has a much higher concentration than the E gene, as occurs in dilutions equal to or greater than 1 × 10−7, the amplification curve of gene E flattens out due to the depletion of reagents in the final cycles of the PCR reaction.
This same flattening effect can be observed for the RNase P curves in reactions in which the E gene has a high concentration of RNA; its amplification can even be inhibited, as shown in the first dilution series of the duplex PCR in Table 1.
Table 2 shows the final reproducibility test between laboratories, for which purpose two extractions of the virus isolate were performed with a concentration of 4.2 × 106 PFU/mL, with concordant results obtained in the duplex PCR E plus RNase P (P = .27).
Reproducibility assay Ct values from two independent dilutions of RNA from virus isolate.
| Extraction 1: Duplex | Extraction 2: Duplex | |||||||
|---|---|---|---|---|---|---|---|---|
| E | RNase P | E | RNase P | |||||
| DILUTION | Ct | Average | Ct | Average | Ct | Average | Ct | Average |
| 1 × 10−1 | 12.4 | 12.6 | 28.3 | 28.4 | 12.6 | 12.6 | 28.3 | 28.3 |
| 12.7 | 28.4 | 12.6 | 28.2 | |||||
| 1 × 10−2 | 15.8 | 15.7 | 27.9 | 27.7 | 15.8 | 15.8 | 27.6 | 27.7 |
| 15.7 | 27.5 | 15.8 | 27.9 | |||||
| 1 × 10−3 | 19.2 | 19.2 | 27.1 | 27.2 | 19.1 | 19.1 | 27.4 | 27.4 |
| 19.1 | 27.3 | 19.2 | 27.4 | |||||
| 1 × 10−4 | 22.5 | 22.4 | 27.6 | 27.5 | 22.5 | 22.6 | 27.8 | 27.7 |
| 22.3 | 27.5 | 22.6 | 27.6 | |||||
| 1 × 10−5 | 26.0 | 25.8 | 28.2 | 28.1 | 25.8 | 25.7 | 28.2 | 28.1 |
| 25.5 | 28.0 | 25.6 | 28.0 | |||||
| 1 × 10−6 | 29.8 | 29.1 | 28.1 | 28.1 | 28.4 | 28.5 | 28.2 | 28.2 |
| 28.3 | 28.1 | 28.6 | 28.1 | |||||
| 1 × 10−7 | 35.6 | 32.9 | 28.1 | 28.1 | 33.6 | 32.4 | 28.1 | 28.1 |
| 30.3 | 28.1 | 31.2 | 28.2 | |||||
| 1 × 10−8 | 36.1 | 38.1 | No data | 28.2 | 0 | 0 | 28.2 | 28.2 |
| 40.2 | 28.2 | 0 | 28.1 | |||||
| NTC | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Contrary to what was reported by the Charité-Berlin protocol, our results show a better sensitivity for the Sarbeco E marker compared to RdRp. To demonstrate the adequate performance of the duplex technique in clinical specimens, 88 RNA samples with a previous result for COVID-19 (39 positive and 49 negative for SARS-CoV-2) were selected (Table 3).
Comparison of Ct values obtained from monoplex and duplex PCR for clinical samples.
| Monoplex | Duplex | Monoplex | ||
|---|---|---|---|---|
| Code | Sarbeco E | E | (RNase P) | RdRp |
| 1 | 21 | 23 | 22 | 25 |
| 2 | 0 | 0 | 29.8 | 0 |
| 3 | 30 | 30 | 30 | 32 |
| 4 | 0 | 0 | 30 | 0 |
| 5 | 0 | 0 | 31 | 0 |
| 6 | 0 | 0 | 31 | 0 |
| 7 | 0 | 0 | 33 | 0 |
| 8 | 0 | 0 | 26 | 0 |
| 9 | 0 | 0 | 23 | 0 |
| 10 | 0 | 0 | 31 | 0 |
| 11 | 23 | 23 | 0 | 24.8 |
| 12 | 26.7 | 26.4 | 27.6 | 28.5 |
| 13 | 23.7 | 23.8 | 33.2 | 25.1 |
| 14 | 19.5 | 19.4 | 26.8 | 21.5 |
| 15 | 0 | 0 | 29.1 | 0 |
| 16 | 0 | 0 | 31 | 0 |
| 17 | 0 | 0 | 32.8 | 0 |
| 18 | 0 | 0 | 30.5 | 0 |
| 19 | 0 | 0 | 22.6 | 0 |
| 20 | 0 | 0 | 27.5 | 0 |
| 21 | 0 | 0 | 26.6 | 0 |
| 22 | 0 | 0 | 25 | 0 |
| 23 | 0 | 0 | 27.8 | 0 |
| 24 | 0 | 0 | 25.5 | 0 |
| 25 | 0 | 0 | 26.7 | 0 |
| 26 | 18.2 | 18.7 | 27 | 25.6 |
| 27 | 25.8 | 25.4 | 26.7 | 29 |
| 28 | 26.5 | 26.8 | 31.3 | 31.4 |
| 29 | 20.2 | 20.7 | 31.9 | 23.4 |
| 30 | 23.8 | 23.5 | 33.9 | 26.5 |
| 31 | 27.8 | 28.1 | 32.5 | 31.2 |
| 32 | 21.8 | 21.8 | 0 | 24.9 |
| 33 | 17.7 | 17 | 33.1 | 17.6 |
| 34 | 20.5 | 20.8 | 26 | 24.3 |
| 35 | 20.2 | 20.2 | 0 | 24.3 |
| 36 | 25.6 | 26 | 32.5 | 29.8 |
| 37 | 17.2 | 17.3 | 24.6 | 17.6 |
| 38 | 25.2 | 31 | 23.6 | 30.4 |
| 39 | 0 | 0 | 25.6 | 0 |
| 40 | 0 | 0 | 26.5 | 0 |
| 41 | 0 | 0 | 27.1 | 0 |
| 42 | 21.1 | 21.3 | 24.3 | 22.3 |
| 43 | 26.2 | 28.7 | 25.7 | 28.8 |
| 44 | 26.2 | 27 | 26.7 | 28.5 |
| 45 | 26 | 25.9 | 27.8 | 25.8 |
| 46 | 27.6 | 27.9 | 28.4 | 26.8 |
| 47 | 31.5 | 35 | 29.9 | 0 |
| 48 | 0 | 36 | 29.9 | 0 |
| 49 | 0 | 36.2 | 29.4 | 0 |
| 50 | 24.2 | 24.7 | 27 | 24.8 |
| 51 | 21 | 21.6 | 26.4 | 22.2 |
| 52 | 0 | 0 | 27.4 | 0 |
| 53 | 0 | 36.2 | 25.7 | 0 |
| 54 | 33 | 32 | 25.8 | 0 |
| 55 | 0 | 0 | 28.3 | 0 |
| 56 | 31.1 | 30.6 | 28.6 | 34.4 |
| 57 | 32 | 32.6 | 31.3 | 34.4 |
| 58 | 31 | 32.4 | 25.2 | 33.2 |
| 59 | 31.9 | 33.9 | 27.3 | 34.7 |
| 60 | 31.4 | 36.3 | 26 | 33.8 |
| 61 | 32.3 | 33.7 | 31.6 | 33.9 |
| 62 | 30 | 32.4 | 26.3 | 32.7 |
| 63 | 33.2 | 33.6 | 27.1 | 34 |
| 64 | 36.7 | 27.9 | 28.4 | 0 |
| 65 | 0 | 35.2 | 29.9 | 0 |
| 66 | 34.1 | 0 | 25.9 | 0 |
| 67 | 37.8 | 36.2 | 30.6 | 0 |
| 68 | 0 | 34.1 | 31.7 | 0 |
| 69 | 0 | 0 | 30.6 | 0 |
| 70 | 0 | 0 | 32.6 | 0 |
| 71 | 20.8 | 21 | 32.2 | 26.8 |
| 72 | 34.7 | 0 | 36 | 0 |
| 73 | 14.2 | 14.4 | 29.1 | 20.5 |
| 74 | 0 | 0 | 26.5 | 0 |
| 75 | 36.3 | 0 | 28.3 | 0 |
| 76 | 0 | 0 | 30.6 | 0 |
| 77 | 0 | 0 | 32.3 | 0 |
| 78 | 37.6 | 0 | 30.7 | 0 |
| 79 | 0 | 0 | 30.2 | 0 |
| 80 | 35.4 | 35 | 30.4 | 0 |
| 81 | 37.1 | 0 | 32 | 0 |
| 82 | 37.3 | 37.8 | 25.3 | 0 |
| 83 | 0 | 0 | 30.1 | 0 |
| 84 | 0 | 0 | 29.8 | 0 |
| 85 | 0 | 0 | 30.6 | 0 |
| 86 | 32.4 | 32.3 | 32.6 | 37.5 |
| 87 | 23.5 | 23.5 | 31.2 | 29.1 |
| 88 | 22.3 | 22.3 | 29.1 | 28.2 |
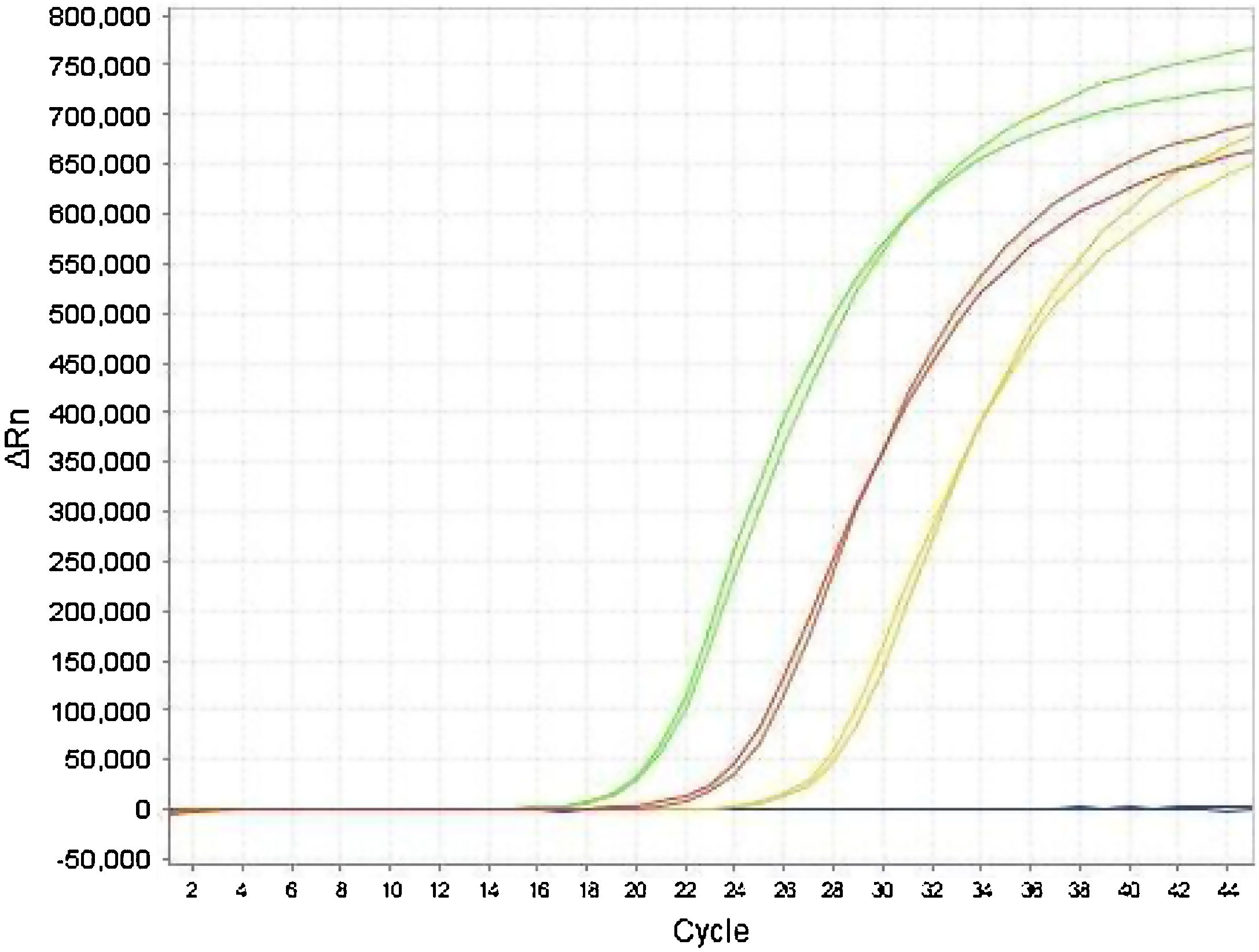
The Ct values of the E gene obtained by monoplex and duplex PCR (E plus RNase P) did not show significant differences in the paired t-test (P = .84). Adequate agreement was observed between the results obtained for the E gene, both in duplex and monoplex (Fig. 2) in clinical specimens, demonstrating that the duplex technique is a good screening test with high sensitivity in the detection of positive samples, which were later confirmed with the specific RdRp gene.
The analysis of agreement between the results of the monoplex E and duplex (E plus RNase P) PCR showed similarity between the two techniques, with a Cohen's kappa coefficient of 0.89, allowing us to state that the strength of the agreement between the two techniques is excellent8, with the rate of the agreement of results between the methods being 89%.
We also calculated the sensitivity and specificity of the duplex PCR (E plus RNAse P) using the monoplex E PCR as the standard, obtaining a sensitivity of 90% and a specificity of 87%. The divergence detected between the two strategies involved samples with Ct >34, corresponding to samples with a low viral load.
DiscussionThe duplex PCR amplification of the Sarbeco E gene published in the Charité-Berlin protocol, along with the RNase P gene, as demonstrated in this article, is an extremely useful tool for SARS-CoV-2 screening in clinical samples, as it optimises the use of reagents, the speed in obtaining results, it improves the installed capacity necessary for diagnosis and thus reduces the cost of the test.
Our results show that duplex PCR does not affect the sensitivity and specificity reported by the Charité-Berlin protocol and they confirm that samples with high viral load (Ct for the E gene below 34) show 100% agreement between the E gene and RdRp.
The analysis of the amplification curves of samples with low viral load showed a decrease in the intensity of fluorescence of the curve of the specific viral gene, which was more marked when there were high concentrations of human RNA. That may be due to the fact that there is a lower likelihood of pairing between complementary sequences (primers and probes) with the viral RNA at lower concentrations, or even the presence of inhibitors in the samples that affect the efficiency of the PCR. In the specific case of the duplex PCR technique, this phenomenon is accentuated by the early amplification of the reaction internal control and the increase in the RNase P fragments available in the reaction, which we suppose induced the flattening observed in the E gene amplification curve and altered its sigmoidal appearance, generating linear amplifications, albeit without modifying the Ct values.
In fact, samples with low viral load (Ct >34), such as samples 48, 49, 66, 75, reported in Table 3, coincide with the Ct obtained from the virus isolate at dilutions greater than 1 × 10−8 (4.2 × 10−2 PFU/mL) in Table 1, where lower reproducibility was found in the PCR results for the five replicates, both for the E gene in monoplex and for the duplex PCR. It is important to highlight that it is the samples with Ct >34 that affect the agreement between the monoplex and duplex PCR, possibly due to being close to the Berlin protocol limit of detection using the Sarbeco E primer, suggesting that the discordant results for these samples could be attributed to the sensitivity of the Berlin protocol and not to the duplex PCR strategy.
Contrary to what was reported by the Charité-Berlín protocol, our results show a better sensitivity for the Sarbeco E marker compared to RdRp. This is in line with the reports of other authors9–11 and shows the need to use another more sensitive and stable confirmatory marker with a low rate of mutations that might affect the efficiency of the PCR. Although the degree to which individuals with a low viral load (Ct >33) are infectious is known to be close to zero12,13, the Ct can vary depending on the quality of the sample. However, from an epidemiological point of view, it is important to know the real number of individuals with the infection in our setting. Therefore, in the diagnosis of SARS-CoV-2, a detailed analysis of the amplification curves is important and, where there is a subtle rise with Ct values above 34 for the E gene, a confirmatory PCR should be performed, ideally directed against a viral gene other than RdRp, which is less sensitive, in order to verify the presence of SARS-CoV-2.
Table 3 shows samples that were amplified with the monoplex and duplex PCR but were negative with the RdRp marker. The moderate specificity of the monoplex and the duplex against RdRp can be explained by the non-specific detection of the Sarbeco E gene, but also by the low sensitivity of the RdRp marker; this may be due to design errors in the primers10 and/or new virus mutations in the Colombian population11.
Finally, the recommendation is for SARS-CoV-2 diagnostic laboratories to have different protocols available and implemented for the diagnosis of the virus which can serve as a back-up for confirming patients with low viral load or mutations in the genome, both of which sometimes lead to inconclusive results by means of a single method. In cases where the results of the different methods differ, we suggest reporting the presence of SARS-CoV-2 as “indeterminate” and repeating the test on a new sample taken 48 h after the original.
Sources of fundingThis work was financed with resources from Colombia's general awards system (BPIN 2020000100152, BPIN 2020000100131) and from the University of Antioquia (2020-36870 and sustainability).
Conflicts of interestThe authors declare that they have no conflicts of interest.
Please cite this article as: Palacio Rua K, García Correa JF, Aguilar-Jiménez W, Afanador Ayala C, Rugeles MT, Zuluaga AF. Validación de una técnica de PCR dúplex usando el gen E y RNasa P para el diagnóstico de SARS-CoV-2. Enferm Infecc Microbiol Clin. 2022;40:428–435.