The incidence of glycopeptide-resistant Enterococcus faecium (GRE) as a nosocomial pathogen has been increasing in recent years in Spain, due to its high capacity to colonise patients and staff for long periods, as well as to persist in the hospital environment, where it can survive for a long time. The main epidemiological presentation of GRE is in the form of sporadic outbreaks,1 such as the one that occurred in the Neonatal Intensive Care Unit (NICU) of our hospital in 2015, and which we discuss below.
From 21 April to 23 June 2015, GRE was isolated in the surveillance rectal swabs (SRS) of 10 neonates admitted to the NICU of our hospital (third level university hospital). The Level IIIA Neonatal Unit (which does not include surgery), consists of six intensive care stations (CIV) and 11 intermediate care stations (CIM) in separate rooms. The personnel responsible for the unit include five on-duty neonatologists plus two on call, a morning shift of four nurses and three auxiliary nurses, and afternoon and night shifts of three nurses and two auxiliary nurses. In 2015, there were 282 admissions with an average stay of 9.10 days, a total recorded use of vancomycin of only 6.5g for the whole year (2.0g in 2014) and no recorded use of ceftazidime in either 2014 or 2015. Newborns are subject to surveillance cultures at the time of admission (nasal, pharyngeal and ear swabs cultured on blood agar for 48h at 35°C in an atmosphere with 5% CO2), and surveillance cultures for extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae, by culture of SRS on chromID-ESBL® plates (bioMérieux), are taken weekly. Following the isolation of GRE, a selective chromogenic plate was added to screen for GRE: chromID-VRE® (bioMérieux). Both plates are incubated aerobically for 48h at 37°C.2
The first GRE isolates were detected in the SRS on 21 April in two patients (colonisation rate [CR]: 33%, 2 patients of 6 admitted). Both GRE were resistant to ampicillin and fluoroquinolones, had high-level resistance to gentamicin and streptomycin, and were resistant to vancomycin and teicoplanin with MIC >16mg/l, but were still susceptible to linezolid. When these results were notified on Friday, 24 April (23 holiday), contact isolation and cleaning precautions were established twice a day with SANIT P20® (4.5% didecyldimethylammonium chloride and 8% monoethanolamine), taking further SRS again that same day, which showed the colonisation of a third neonate (CR 30%, 3 of 10). In the weekly surveillance of 28 April, another four neonates had SRS with growth of GRE with the same susceptibility phenotype, making a total of six colonised neonates remaining admitted at that time (one of the three previous ones had been discharged) (CR 55%, 6 of 11). In view of this situation and the clinical suspicion of sepsis in one of the colonised patients, on 29 April, a multidisciplinary committee decided not to accept any new admissions throughout the Unit, to put the isolation into effect in a cohort, situating all the colonised patients in the CIM room with exclusive staff (not shared with the CIV room), reinforce the team of cleaning staff and compliance with cleaning protocols, reinforce contact isolation measures (education of staff and parents) and restrict parent visits, providing them with personalised lockers for personal items and use of different toilets.
On 4 May, another neonate was found to be colonised (CR 43%, 3 of 7). On 14 May, after the discharge of the last colonised child, we proceeded to carry out a double terminal cleaning following the cleaning protocols for critical areas of our hospital: removal of the nursing material and thorough cleaning (SANIT P20®) including walls and ceilings, drip-stand feet, bed in its entirety and mattress, rubbish bins and side tables, twice. After the completion of the cleaning, environmental samples were taken, including different surfaces, medical equipment, taps, incubators and cradles, dressing trolleys, scales, computer keyboards, etc., using swabs pre-moistened in brain-heart infusion broth (BHI-T®bioMérieux) which were introduced in the same medium; then, after shaking, 0.5ml was inoculated on a plate with selective medium chromID-VRE. After 18–24h of incubation of the broth (37°C, aerobically), 0.5ml was re-spread in the chromID-VRE3 medium. GRE was isolated in a mobile column (support equipment).
After the cleaning, before receiving the results of the cultures, the Unit was reopened due to pressure of care and, in the SRS of 19 May, GRE was isolated in a new patient (CR 20%, 1 of 3). That patient was discharged on 20 May along with another patient, leaving the CIM room empty (only one child remained in CIV). After terminal cleaning of CIM, environmental samples were taken, isolating the same GRE in the child's cot. A further terminal cleaning was carried out, with the CIM reopening on 25 May. After a month with no cases detected, a new carrier was detected once more on 23 June (CR 20%, 1 out of 5). Since then, and to date, no new isolates have been documented.
To sum up, the outbreak affected five boys and five girls, with gestational ages between 29 and 40 weeks, with adequate weights for gestational age and an average stay of 18.3 days (range 5–40 days). The date of admission was the date of birth, except for three infants who were admitted at 48h, 7 and 9 days of life. None of the patients had been treated with vancomycin. In terms of symptoms, none of the children had diarrhoea, and only one had clinical sepsis with negative blood cultures and no GRE isolated in any other clinical sample which was effectively treated with linezolid for seven days. Six neonates had prior negative rectal swab cultures, one had a previous rectal swab positive for VIM-type carbapenemase-producing Enterobacter cloacae and the other three had no previous SRS, although they had been admitted to the NICU the day they were born and culture of their neonatal pharyngeal, nasal and ear swabs showed no growth.
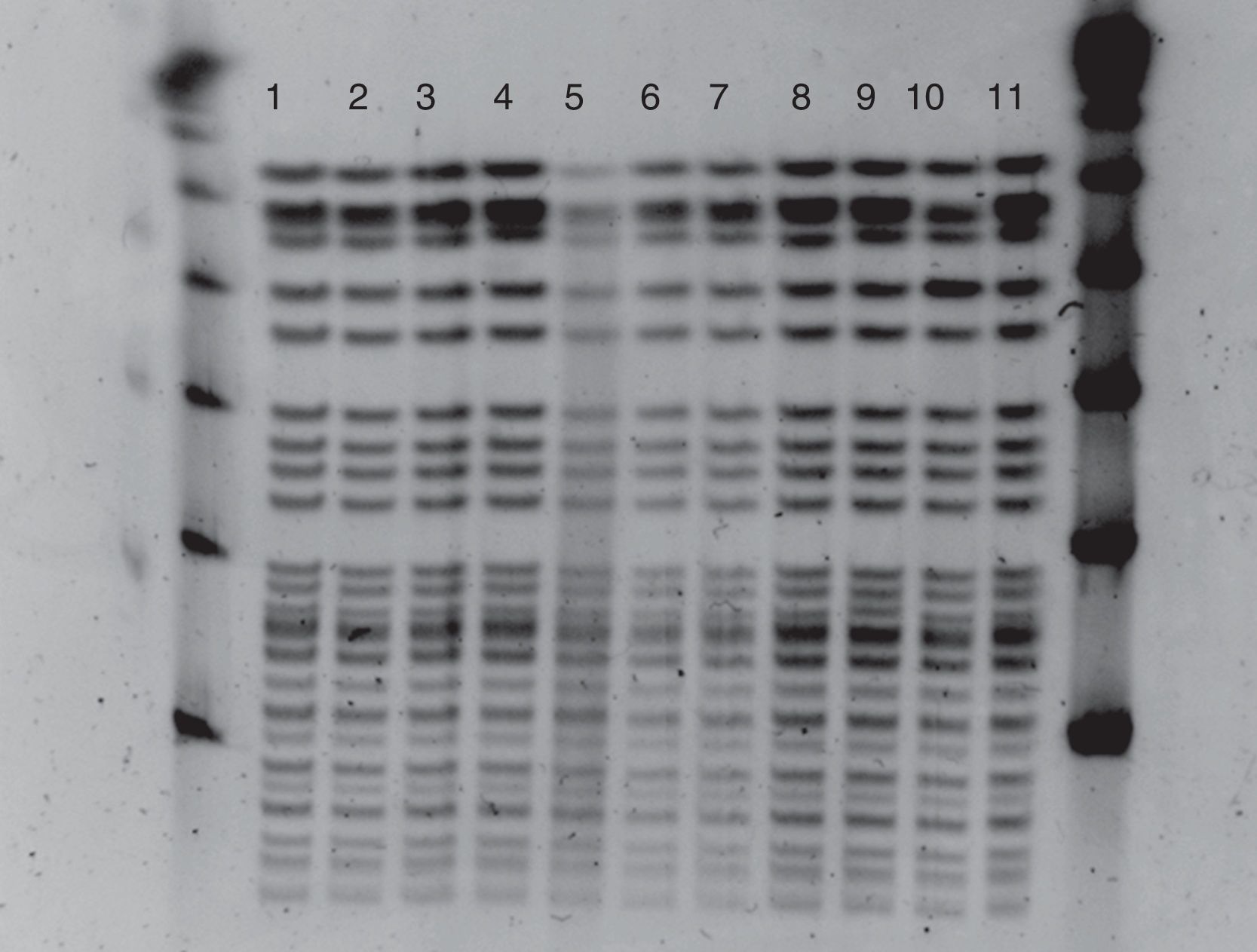
The identification and susceptibility study was carried out by VITEK2® (bioMérieux), confirming resistance to glycopeptides by E-test (bioMérieux). The strains were sent to the Antibiotic Resistance Surveillance Programme of the Centro Nacional de Microbiología [National Microbiology Centre] for study of the mechanism of resistance and molecular epidemiology. The vanA gene was detected in all of them. Pulsed-field gel electrophoresis (PFGE), after digestion of the total DNA with the SmaI restriction enzyme, showed that the isolates (including the two environmental isolates) had the same profile (Fig. 1) belonging to the sequence type 17 (ST17), identified by multilocus sequence typing (MLST).
ST17 gives its name to the clonal complex 17 (CC17) widely distributed in the hospital environment all over the world.4 In addition to the classic resistance to ampicillin and fluoroquinolones of CC17, outbreaks produced by isolates of this clonal complex resistant to glycopeptides have increased in recent years.4,5
In our case, routine surveillance allowed rapid detection of the outbreak in which ten patients were colonised.6,7 The environmental cultures demonstrated the high capacity GRE has to disseminate, as well as its persistence on inanimate surfaces.8
The first measures of contact isolation taken after the first two cases were not enough to prevent the rapid colonisation of another five children, perhaps influenced by the one-day delay in implementing the measures, combined with the weekend (Thursday holiday in our region). We wish to highlight the difficulty, in terms of the success of the implemented measures, of not having individual rooms and the patient areas being very close to one another. Moreover, due to the pressure of care, the Unit was reopened immediately after the terminal cleaning, before we had the results of the first environmental cultures (in which GRE was isolated), and that obviously made it possible for new cases of colonisation to occur.
In this study, it is also worth mentioning that the chromID-ESBL® plates (non-selective for GRE) allowed the detection of GRE. Therefore, although the chromID-VRE plate facilitated the detection of GRE during the outbreak, in the absence of outbreaks, we could stop using it to reduce costs. We are aware of how time consuming surveillance cultures are, and of the fact that they are very often not cost-effective. However, in critical patients such as neonates, who are predisposed to infections, they have been shown to be effective in presuming the causative agents of infection,9 and, in view of the therapeutic challenge posed by such problems in neonates, they can enable the timely initiation of adequate antimicrobial therapy.10 Moreover, the early detection of colonised patients can help to improve infection control measures, which should limit rapid and extensive spread and avoid a potential increase in infections among the patients (colonisation–infection ratio, 10:1).1,6
In conclusion, we have described an outbreak of GRE-ST17 in which performing surveillance studies for carriers of multidrug-resistant bacteria enabled us to detect the outbreak early and quickly apply control measures, which included contact isolation, patient cohorts, reinforcing of cleaning protocols, and temporary closure of the unit to new admissions.
To Elena Infante López (specialist physician, Neonatal Intensive Care Unit), and Aurora Sacristán Salgado (specialist physician, Preventive Medicine Service) of the Hospital Universitario Río Hortega (HURH), for their collaboration during the outbreak and support in the preparation of this text, and to Soledad Sañudo García (physician, Admission and Clinical Documentation) and Tomás Caro-Patón Carmona (specialist pharmacist, Hospital Pharmacy Department) also of the HURH, for the data provided.
Please cite this article as: López-Urrutia L, de Frutos M, Oteo J, Eiros JM. Brote por Enterococcus faecium ST17 resistente a glucopéptidos en una Unidad de Neonatología. Enferm Infecc Microbiol Clin. 2018;36:198–200.