In molecular biology, the conservation of genetic material from clinical samples is key when performing experiments with RNA. Therefore, it is important to take the maximum precautions to avoid its degradation, mainly due to RNases that can cause fragmentation of genomic material. These are robust proteins with enzymatic activity that participate in various physiological processes and their main function is to hydrolyse and degrade RNA, and they can also be introduced exogenously into experiments.1,2
When clinical samples are kept in the laboratory for long periods of time, their RNA is especially vulnerable to degradation,3,4 which could influence viral diagnosis by decreasing the sensitivity of amplification techniques.
Various studies have shown that ribonuclease inhibitors exhibit a wide spectrum of inhibitory activities against RNases, effectively protecting RNA during laboratory procedures, allowing great flexibility in experimental design. Their use can be a solution to minimize these degradation problems.5–7
The objective of this study was to evaluate the integrity of viral RNA from clinical samples after storage at 4°C, after a standard working time period (10 days), which is considered sufficient to carry out all the routine processes involved in the viral diagnosis of a sample, and to analyze the efficacy and performance of an RNase inhibitor to prevent its degradation.
Of the samples processed in the Virology Unit of the Hospital Central de Asturias [Central Hospital of Asturias] for two months, 67 exudates (42 nasopharyngeal and 25 pharyngeal) collected in a viral transport medium were studied. Extraction and purification of the genomic material was carried out with the MagNA Pure LC Total Nucleic Acid Isolation Kit Reagent, in the MagNA Pure LC 2.0 automated system (Roche Diagnostics, Switzerland). 100μl was obtained for a subsequent amplification of fragments of the viral genome and laboratory syndromic diagnostic protocols were applied. In all of them, the quantity and quality of the sample was evaluated, and with the amplification of the human β-globin gene, the normalized viral load (by number of cells) was reported. From the total nucleic extract, two aliquots of 10μl were separated and in one 1μl RNase® Inhibitor (Applied Biosystems, USA) was added. Both were stored at 4°C.
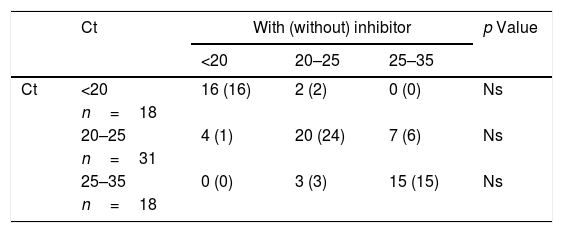
The 67 samples included underwent the usual laboratory diagnostic process for enterovirus (n: 53), parainfluenza virus (n: 11), and influenza A virus (n: 3). After 10 days, the two stored aliquots underwent the same PCR and the amplification cycles (Ct) were compared with and without inhibitor. They were also divided into three groups according to their Ct (<20, 20–25 and 25–35 cycles) and type of virus.
All statistical calculations were performed with the computer program, GraphPad InStat v2.04a (GraphPad Software, USA) according to the specific requirements.
In Table 1, we observe that the presence of viral RNA is evident in all the samples studied, and no statistically significant differences were found in any group after 10 days regardless of whether or not an inhibitor was added. Furthermore, the mean Ct does not vary significantly in any virus.
Ct results in each of the protocols and their mean according to the type of virus.
| Ct | With (without) inhibitor | p Value | |||
|---|---|---|---|---|---|
| <20 | 20–25 | 25–35 | |||
| Ct | <20 | 16 (16) | 2 (2) | 0 (0) | Ns |
| n=18 | |||||
| 20–25 | 4 (1) | 20 (24) | 7 (6) | Ns | |
| n=31 | |||||
| 25–35 | 0 (0) | 3 (3) | 15 (15) | Ns | |
| n=18 | |||||
| Total | Standard | With inhibitor | Without inhibitor | ||
|---|---|---|---|---|---|
| 24.78±3.44 | 25.07±2.37 | 25.28±2.44 | |||
| (21–28) | (22–27) | (22–27) | |||
| Virus | IA | 28.67±4.16 | 28.34±3.78 | 28.67±5.13 | Ns |
| n=3 | (24–32) | (24–31) | (23–33) | ||
| PIV | 23.54±5.80 | 24.09±5.75 | 24.18±5.87 | Ns | |
| n=11 | (15–30) | (17–31) | (17–32) | ||
| ETV | 22.13±4.12 | 22.79±5.53 | 23.00±5.25 | Ns | |
| n=53 | (12–29) | (11–36) | (12–34) |
Ct: amplification cycles; ETV: enterovirus; IA: Influenza A virus; Ns: not significant; PIV: Parainfluenza virus.
The samples repeated the amplification range in 59 (88.1%) of the occasions after 10 days, and in 58 (86.6%) of the cases, when RNasin® was added to the extracted sample.
This study shows that, although the use of RNasin® is usually recommended in RNA virus management protocols, it does not seem essential, at least in the viruses studied. In some cases, the performance in the samples with and without an inhibitor decreased slightly, and it is important to note that no sample was undetectable after 10 days.
Thus, we maintain that for viral studies that are conducted in a period of 10 days stored at 4°C it does not decrease the diagnostic capacity and it would not be necessary to add preservative reagents, which would decrease the handling of the samples and also would not imply an increase in the cost of the diagnostic tests. It would be useful to conduct studies over longer periods of time to establish from what time possible degradation of RNA could be minimized by using an enzyme inhibitor.
Please cite this article as: García Fernández X, Álvarez-Argüelles ME, Rojo S, de-Oña M. Estabilidad del ARN viral en muestras clínicas para el diagnóstico viral. Enferm Infecc Microbiol Clin. 2020;38:297–298.