Tuberculosis is still a serious public health problem, with 10.8 million new cases and 1.8 million deaths worldwide in 2015. The diversity among members of the Mycobacterium tuberculosis complex, the causal agent of tuberculosis, is conducive to the design of different methods for rapid diagnosis. Mutations in the genes involved in resistance mechanisms enable the bacteria to elude the treatment. We have reviewed the methods for the rapid diagnosis of M. tuberculosis complex and the detection of susceptibility to drugs, both of which are necessary to prevent the onset of new resistance and to establish early, appropriate treatment.
La tuberculosis continúa siendo hoy en día un grave problema de salud pública con 10,8 millones de casos nuevos y 1,8 millones de muertes a nivel mundial en el año 2015. La diversidad existente entre los miembros del complejo Mycobacterium tuberculosis que producen la tuberculosis permiten el diseño de métodos para su rápido diagnóstico. Así mismo, las mutaciones en los genes implicados en los mecanismos de resistencia permiten escapar a la bacteria del tratamiento. Hemos revisado los métodos de diagnóstico rápido de M. tuberculosis complex y de la detección de la sensibilidad/resistencia a los fármacos; ambas cosas son necesarias para detener la aparición de nuevas resistencias e instaurar un tratamiento precoz y correcto.
The first indications of tuberculosis were found in mummies from 5000 BC and, although 135 years have passed since Robert Koch discovered the bacillus that causes “white phthisis”, tuberculosis (TB) continues to be a serious worldwide public health problem today. According to the latest data reported by the WHO, in 2015 there were 10.8 million new cases and 1.8 million deaths worldwide from this infectious respiratory disease.1 In that same year in Spain, 4191 new cases were reported (12/100,000 inhabitants).
Since the discovery of the first drugs, streptomycin (SM) in 1943, isoniazid (INH) in 1952 and rifampicin (Rif) in 1959, resistant strains began to appear, making it necessary to treat the disease with a combination of drugs. Insufficient or disrupted treatment causes the appearance of strains resistant to one or various drugs, leading to the appearance of multidrug-resistant strains (MDR), defined as those resistant to at least INH and Rif. When resistances are not identified prior to the start of treatment, the situation becomes particularly dangerous, since patients infected with monoresistant strains have a high risk of developing additional resistances to other drugs if they receive standard therapy. A poor administration of second-line drugs, which are less effective and have more serious side effects, can lead to extensively drug-resistant (XDR) strains, which are MDR strains with additional mechanisms of resistance to at least one of the 3 injectable drugs: kanamycin (KM), amikacin (AK) and capreomycin (CPM), and to any fluoroquinolone used against Mycobacterium tuberculosis (MTB).1 Cases of XDR-TB in practice are nearly impossible to treat. Cases have been reported with XDR strains that are even resistant to delamanid and bedaquiline, antituberculosis drugs introduced recently to treat XDR-TB.2 Therefore, early diagnosis of TB, obtaining rapid susceptibility results, and proper prescribing of drugs are crucial for adequate treatment and control of the disease.
The Mycobacterium genus is the only one in the Mycobacteriaceae family, of the actinomycetales order, characterised by having a high guanine and cytosine content (G+C). It appeared as we know it today about 15,000 years ago. There are more than 170 reported species, most of which belong to the nontuberculous mycobacteria (NTM) group, and only a few, which share 99.9% of similarity in their genomes, that belong to the M. tuberculosis (MTBc) complex, all causing TB in humans or various animal species. MTBc includes various lineages that have evolved genotypically and phenotypically after clonal expansion from a common ancestor, adapting to various hosts.
At the time of a clinical suspicion of TB, the first step is to confirm the diagnosis microbiologically. Bacilloscopy or visualisation of the micro-organisms from a smear of the pathological product stained with Ziehl–Neelsen stain continues to be the fastest, cheapest and most accessible way to diagnose smear-positive cases, although their susceptibility compared to the culture varies between 25 and 90%, depending on the sample being studied and on the bacterial load of said sample, as well as the quality of the observer. Culture in Lowenstein–Jensen and Coletsos solid medium has the drawback of MTBc taking more than 15 days to grow. The first advance in rapid diagnosis was thanks to S. Siddiqi, who designed the Bactec 460TB radiometric system at the end of the 1970s, which reduced the time to obtaining a positive culture and increased sensitivity.3 It also provided the additional advantage of allowing an antibiogram to be conducted, significantly decreasing the time to obtain the result compared to the Canetti proportion method.4 Identification through phenotypic methods: optimal temperature for growth, pigmentation of the colonies and biochemical characteristics, is slow and sometimes complex, particularly in many of the NTM species. In the 1990s, the first genetic identification techniques appeared, again achieving greater speed and accuracy in the mycobacteriological diagnosis.5 The need to provide a rapid diagnosis today is perhaps the most important aspect for the control of tuberculosis. This has driven research and development of new diagnostic methods based on molecular biology in recent decades. Ideally we should have a sensitive, rapid, reasonably cheap technique that does not need complex technical means since most cases occur in underdeveloped countries with limited financial resources.
With this paper we are not attempting to provide an exhaustive description of all the methodologies available or currently in development, for which we refer the interested reader to more specialised literature,6,7 but to review the various existing approaches to rapid and microbiological diagnosis of tuberculosis.
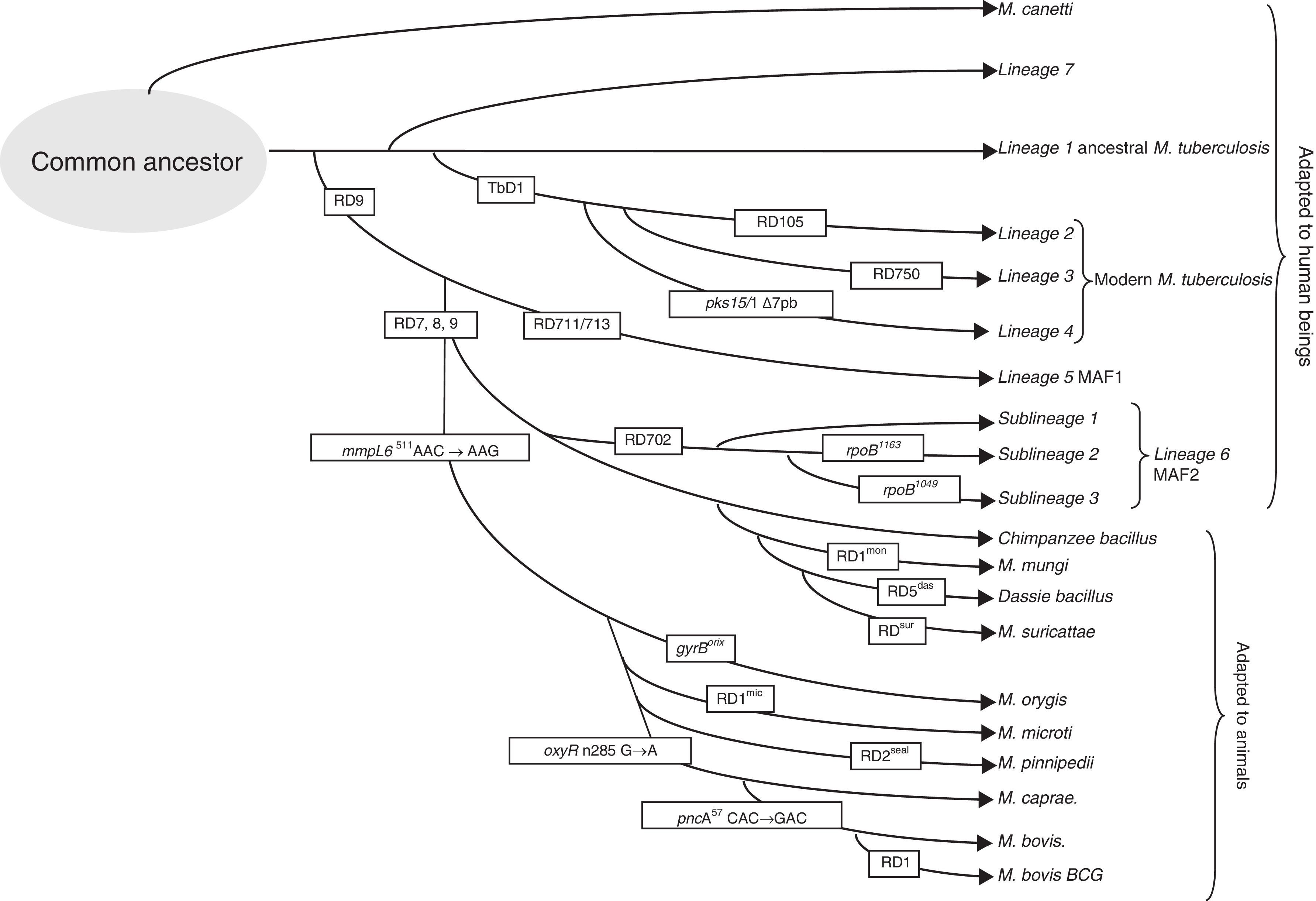
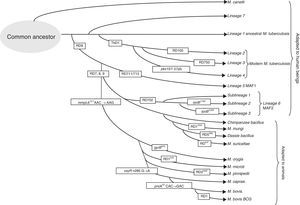
Genomic differences in the members of the M. tuberculosis complexPolymorphisms have been detected in its genomes due to the loss of large sequence polymorphisms (LSP), mutations affecting a nucleotide (SNP) or variations in the repetitive elements in their genomes. The specific deletions in the genome of the different members of the complex have been called differential regions (DRs)8 and can be used to identify them and classify them into 8 different lineages (Fig. 1). MTB includes lineages 1–4 and has adapted selectively to humans. Despite infecting a third of the world's population, it has not established itself in the animal world.12 The presence or absence of the region called TbD1 differentiates between ancestral (L1) and modern MTB.8 Other differences in SNP makes it possible to classify the isolates in sublineages within the strains of modern MTB: L2 that includes the Beijing family, L3 that includes the CAS family, and L4 that includes the Latin American family, the most prevalent in Europe. To differentiate the different lineages and sublineages or families, different techniques have been proposed: multiplex PCR, pyrosequencing, SNAPshot, and gene group sequencing.13–15 The loss of a region called RD9 would separate MTB from Mycobacterium africanum, also associated with human hosts (L5 and L6), and from the rest of lineages from the complex associated with animal hosts, lineage 8.16 A group of strains that circulates among humans with apparently less virulence is related to lineage 6, also called M. africanum, West African 2 (MAF2). These strains have lost regions RD7, RD8, RD9 and RD10, as occurs in strains associated with animals including Mycobacterium bovis, M. bovis bacillus Calmette-Guerin (BCG), Mycobacterium caprae, Mycobacterium oryx, Mycobacterium pinnipedii, and those described more recently, the so-called chimpanzee bacillus, dassie bacillus, Mycobacterium mungi and Mycobacterium suricattae, which have been isolated from chimpanzees rock hyraxes, banded mongooses and meerkats, respectively.17
Rapid microbiological diagnosisThe need for early diagnosis gains special importance if we keep in mind that nearly 20% of transmissions are due to patients with negative bacilloscopy and positive culture,9 and with the increasing growth of isolates with resistance to various drugs.
Despite the fact that microscopy continues to be the most widespread and cheapest method for immediate diagnosis, its drawbacks – which have already been mentioned—have driven research and development of new methods, among which the most used ones, without a doubt, are those based on molecular biology.
BiomarkersA theoretical rapid method to detect active infections is the detection of biomarkers from minimally invasive or non-invasive samples; however, despite some development in this line, to date no techniques are available that can meet the required needs.
Serology. Just like for many infectious diseases, detection of circulating antibodies has been tested for TB. Nevertheless, due to its low sensitivity and cross-reactions, the WHO has formally advised against using it.
Antigen detection. Another possibility is the search for an MTB antigen in patient products. In this respect, there are some tests on the market or being studied.
Detection of lipoarabinomannan in patients’ urine, lipopolysaccharides present in the mycobacterial wall have been developed by Alere (LF-LAM, Alere Determine™ TB LAM Ag, Alere Inc., Waltham, MA, USA). This test is a lateral flow immunochromatography to be conducted at point-of-care that only requires a minimal amount of urine, 25min of incubation, and is very cheap. One disadvantage of this technique is that this lipopolysaccharide is not only found in MTB, but also in other mycobacteria. Both the WHO's report18 and a recent review of the Cochrane Library19 conclude that its sensitivity, both in patients infected with HIV and in their partners, is low, not recommending its use for screening or diagnosis, although it could be useful in patients with less than 50CD4/μl, where its sensitivity increases.
Volatile organic compounds. Detection of different components in patients’ exhaled air constitutes an area of growing interest. Various devices with a point-of-care philosophy are in a development or evaluation phase. The patient is asked to exhale or cough into a device (after nebulisation with physiological saline solution) and the volatile organic compounds or the presence of MTB antigens are analysed. Studies are being conducted with promising results.7
TB-TAM. Another biomarker is the T-cell activation marker (TAM), described in 2014,20 that measures the expression of a membrane protein (CD127) in CD4 T-cells exposed to MTB antigens via flow cytometry. In preliminary trials, a sensitivity of 83% and a specificity of 96.6% were reported. A commercial version of this technique will soon be available.
Detection of M. tuberculosis in clinical samplesIn the last 20 years, intensive work has been done to achieve trustworthy, direct diagnoses based on clinical samples. The multiple techniques that have been put into practice and marketed are a result of this work.
Mycobacteriophages. The use of mycobacteriophages for detection in a direct sample comes from the second half of the 20th century, when mycobacteriophages were discovered. Since then, different types of phages and techniques have been tested. The two most important techniques are: ‘Phague Amplified Biologically Assay’, the first that was used, is based on the phage's capacity to lyse cells of a Mycobacterium smegmatis culture, used as a control. If the patient's sputum contains viable MTB, once the phages have been released by lysis, they replicate and create “holes” in the control agar. The second, more recent technique, ‘Luciferase Reporter Phage Assay’, consists of a phage into which the luciferase gene has been incorporated. The presence of the luciferin substrate, if mycobacteria are in the sample, infects them and causes luminescence that can be measured with a luminometer. These tests are performed in 2–3 days, are simple and cheap and have good specificity, but variable sensitivity that decreases in samples with low bacterial loads.21
Molecular methods. The greatest advances in recent years in rapid diagnosis of infectious diseases, including TB, have, without a doubt, been produced in the field of molecular biology. The different commercial kits are based on genetic polymorphisms that differentiate the lineages with greatest clinical impact: MTB, M. bovis, M. bovis BCG and M. africanum. The sensitivity, reproducibility, speed of the results, increasing automation and decrease in cost have made these methods routine in microbiology laboratories.
The classic tests for nucleic acid amplification, with different techniques, are based on amplification of target sequences and their subsequent detection, whether via chemiluminescence, colorimetry or fluorometry.22 Including:
Amplified M. tuberculosis Direct Test (AMTD, Gen-Probe Inc., USA). It amplifies 16S rRNA using an isothermal reaction and then hybridises it with MTB-specific probes that it detects using chemiluminescence. It was the first test approved by the Food and Drug Administration (FDA) in 1995 for samples with positive bacilloscopy (>90% sensitivity) and, in 2000, for negative bacilloscopy (lower sensitivity, 65–93%). The main disadvantages are the absence of internal control and that it is not automated.
Cobas Amplicor M. tuberculosis test (Roche Diagnostic System Inc., Switzerland). It amplifies 16S rRNA using PCR, hybrid with probes, and performs the detection via colorimetry. It has also had FDA approval since 1996. It has a sensitivity similar to the previous test in positive bacilloscopy, and somewhat less if negative, maintaining very good specificity (91–100%). Unlike AMTD, it can be automated and has internal control.
BD Probe Tec ET Direct TB System (DTB, Becton Dickinson). It amplifies two targets: 16S rRNA and IS6110 using the SDA (strand displacement amplification) technique, which is an isothermal enzymatic reaction, detecting the final product using fluorescence. Its sensitivity is higher than 90% in samples with positive bacilloscopy, and it decreases considerably, with a varying range in different tests, in those with negative bacilloscopy.
TB-LAMP (Eiken Chemical Company Ltd., Japan). This is a test based on a fairly recent technique called LAMP (loop-mediated isothermal amplification), which amplifies DNA very quickly (in less than an hour), with the results read at a glance based on turbidity, colour or fluorescence and with very few infrastructural requirements. Its results are promising, and, in a recent review for sputum samples,23 the WHO concluded that it could be an alternative in areas with few resources.
Other methods that detect the product amplified by solid-phase hybridisation consist of a nitrocellulose strip that has the specific probes affixed to it. These include, relatively recently (2010), the GenoQuick MTB (Hain Lifescience) method, which targets the specific insertion sequence for MTBc, IS6110 and conducts the detection via hybridisation in lateral flow. The technique is completed in 3h, the post-amplification phase is manual and has high sensitivity.
There are 2 other solid-phase hybridisation assays on the market: INNO-LiPA Rif. TB (Innogenetics, Belgium) and GenoType MTBDRplus (Hain Lifescience) that, in addition to identifying MTBc, are able to detect resistance mutations, that we will discuss later on. They are very easy to interpret and can be used both in direct samples and in cultures, both with very high sensitivities, and in both the post-amplification phase can be done manually or automatically.
Real-time PCR. Real-time PCR performs the target amplifications and detections simultaneously using fluorescent TaqMan or Molecular Beacons probes, in general. These probes incorporate a fluorochrome and an inhibitor that are very close together, so that when there is a bond with the amplified target, separation occurs and a light is emitted that can be detected and is proportional to the number of micro-organisms in the sample. The appearance of real-time PCR has constituted a great advance in microbiological diagnosis, including mycobacteria identification, reducing contamination problems by performing the entire test in a single tube that does not need to be handled, increasing sensitivity and having the advantage of obtaining a quantification of the micro-organism studied in the sample.
Cobas Taqman MTB (Roche Diagnostics) is an adaptation of Cobas Amplicor MTB from the same manufacturer, previously seen (it amplifies 16S rRNA), which seems to have increased its specificity with this technique.
Fluorotype MTB (Hain Lifescience), released in 2011, has IS6110 as its target, and can also be used in extra-pulmonary samples and the result is analysed by the programme that evaluates the fluorescence emitted at different temperatures, recognising the one corresponding to MTB. Various studies have recognised a 100% sensitivity in positive bacilloscopy samples and between 55 and 85% in negative bacilloscopy samples.24
Abbott Real Time MTB also uses IS6110 as its target, with equal sensitivity to the previous test in positive bacilloscopy and somewhat higher in negative bacilloscopy, although it is only indicated in respiratory samples. Another test from the same brand is available (Abbott RealTime MTB RIF/INH Resistance), which also detects resistance to INH and Rif.
Anyplex plus MTB/NTM/DR-TB (Seegene Inc., Korea), not only differentiates between MTB and NTM since, if positive, in a second test (taking only 40min), it can detect resistance to INH and Rif.
Finally, Xpert MTB/RIF (Cepheid) should be mentioned, which is a totally automated system, without any handling except for the insertion of the sample in the cartridge, that simultaneously detects the presence of MTB and resistance to Rif. As a disadvantage, it should be noted that this test is more expensive than other systems, although there is a lower-cost agreement for countries with limited resources. In 2011, after an evaluation of its effectiveness, the WHO recommended it as a first diagnostic step in countries with high rates of MDR-TB.25
Identification based on positive culturesRapid identification of strains obtained through cultures is equally important for distinguishing MTB from others. Currently there are two very fast methods not based on molecular amplification:
Immunochromatography. There are various commercial tests based on detecting the MPT64 antigen, which is a protein excreted by nearly all MTBc (except if they have mutations in the mpt64 gene) and except for some M. bovis BCG. The test can be performed directly on a liquid medium, or based on a solid medium using an extraction buffer. The “soap dispenser” type device has affixed anti-MPT64 antibodies and a coloured line appears less than 15min after adding the bacterial suspension. The sensitivity and specificity are very close to 100%.
Mass spectrometry. In recent years, identification of microbiological cultures in general has experienced a real revolution, going from the traditional biochemical methods to mass spectrometry using MALDI-TOF (matrix-assisted laser desorption/ionisation-time of flight). In the field of mycobacteria, this technique has taken somewhat longer to take hold, due to the various non-standard protocols and to the worse results. However, little by little, improvements have occurred, both in the technique and in the systems’ database libraries, which is leading to these being used with increasing frequency, and recent studies indicate that their concordance with genetic methods is very high.26 Surely, these will soon be one of the most widely used methods to identify mycobacteria rapidly and reliably in laboratories that have access to spectrometry.
Molecular techniquesNucleic-acid probes. This was the first nucleic-acid based detection test available. It consists of designing fluorescent DNA probes complementary to the rRNA of the mycobacteria that we wish to identify and, when they bond, a chemiluminescent reaction occurs. It can be used both for liquid and solid media, and its sensitivity and specificity are very good. Genetic amplification is not performed since the RNA is obtained by sonicating the bacterial suspension. Therefore, complex technical means are not needed and the result is obtained in less than 2h.
There are probes for MTBc and for the most significant NTMs in clinic practice such as Mycobacterium avium-intracellulare, Mycobacterium kansasi and Mycobacterium gordonae, and their major disadvantage is that they must be oriented to the mycobacteria in question in order to be able to select the appropriate probe, and if the result is negative, another must be tried.
Genetic amplification. These tests consist of amplifying the nucleic acids of certain genetic regions and then analysing them. The amplification is performed via PCR of the DNA extract obtained from the solid or liquid culture, and the final product can be studied in various ways:
PRA (PCR-RFLP of the hsp65 gene). This is a relatively simple, cheap, rapid technique based on amplification of said gene, whose sequence has some conserved and other variable regions. This means that different species can be differentiated. Once amplified, it undergoes enzymatic digestion and then electrophoresis, analysing the band pattern. The band pattern result is characteristic for the different mycobacteria species.
16S rDNA sequencing. The sequencing of 16S rDNA, after PCR amplification, has revolutionised the bacterial taxonomic system in recent years, allowing for the establishment of phylogenetic relationships between different micro-organisms, giving rise to a new classification system and to the possibility of absolutely accurate identification thereof. It is a complex and expensive technique, therefore its use is limited to specialised laboratories, although faster and cheaper applications are being worked on, such as pyrosequencing, which makes this technique much more accessible.
Solid-phase hybridisation. As we previously mentioned, solid-phase hybridisation techniques subject the product of genetic amplification to nitrocellulose strips to which specifically marked probes are attached. After development via enzymatic reaction, we obtain a pattern of bands that indicates which mycobacterial species it corresponds to. There are two commercial systems on the market: INNO-LIPA MYCOBACTERIA (Innogenetics, Belgium) and Geno-Type (GT) Mycobacterium (Hain Lifescience, Germany). In the former, amplification is performed on the 16S-23S intergenic spacer, and the nitrocellulose strip has 22 probes that identify the MTB complex and 16 other clinically-relevant NTMs. The GT Mycobacterium amplifies the 23SrRNA gene and there are 3 different formats. The GT MTBC identifies the subspecies of MTBc, the GT CM identifies MTBc and 13 species of NTM and the GS AS identifies 16 more species. Although these assays are validated for testing strains, it is possible to perform them on direct samples with positive bacilloscopy.
There are also microarray solid-phase hybridisation techniques that allow for a greater number of probes, but currently they are not sufficiently developed for marketing in clinical practice.
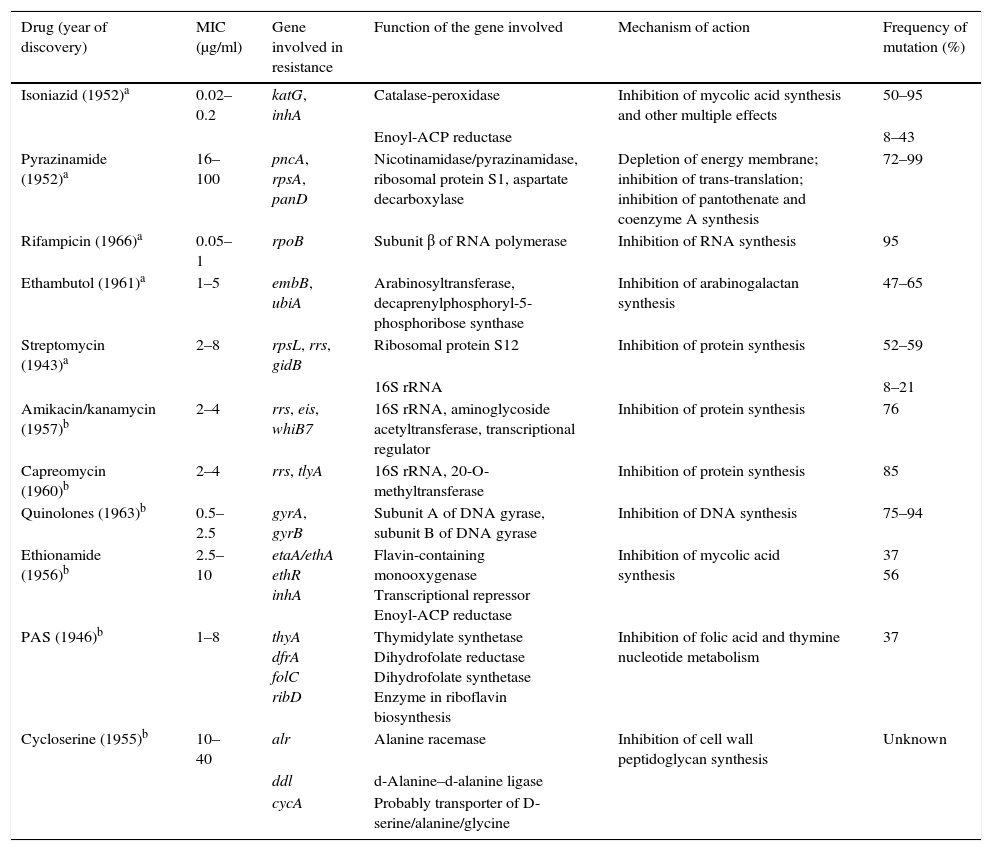
Study of mechanisms that confer resistanceThere are some methods to determine the susceptibility of mycobacteria that are faster than the traditional proportional method and that are recommended by the WHO in places where the necessary technology may not be available, such as direct microscopic observation drug susceptibility (MODS), tests with colourimetric redox indicator (CRI) or nitrate reductase assay (NRA). However, the greatest push in this field has been, without a doubt, the development of the detection of resistance mechanisms using molecular techniques universally recommended by the WHO. In this section, we summarise the molecular events that have been selected in the evolution of clinical strains of MTB for acquisition of resistance over the course of infections and outbreaks. The de novo drug resistance mechanism among MTBc members is due to acquiring individual mutations, in contrast to other bacteria in which the resistance acquisition mechanism is mainly due to horizontal transfer of cassettes with resistance genes. For several years, work has been ongoing to determine the mutations that cause resistance to antituberculosis drugs. Table 1 summarises the genetic bases of MTBc resistance to first- and second-line drugs.
Resistance mechanisms in Mycobacterium tuberculosis against first- and second-line drugs.
| Drug (year of discovery) | MIC (μg/ml) | Gene involved in resistance | Function of the gene involved | Mechanism of action | Frequency of mutation (%) |
|---|---|---|---|---|---|
| Isoniazid (1952)a | 0.02–0.2 | katG, inhA | Catalase-peroxidase | Inhibition of mycolic acid synthesis and other multiple effects | 50–95 |
| Enoyl-ACP reductase | 8–43 | ||||
| Pyrazinamide (1952)a | 16–100 | pncA, rpsA, panD | Nicotinamidase/pyrazinamidase, ribosomal protein S1, aspartate decarboxylase | Depletion of energy membrane; inhibition of trans-translation; inhibition of pantothenate and coenzyme A synthesis | 72–99 |
| Rifampicin (1966)a | 0.05–1 | rpoB | Subunit β of RNA polymerase | Inhibition of RNA synthesis | 95 |
| Ethambutol (1961)a | 1–5 | embB, ubiA | Arabinosyltransferase, decaprenylphosphoryl-5-phosphoribose synthase | Inhibition of arabinogalactan synthesis | 47–65 |
| Streptomycin (1943)a | 2–8 | rpsL, rrs, gidB | Ribosomal protein S12 | Inhibition of protein synthesis | 52–59 |
| 16S rRNA | 8–21 | ||||
| Amikacin/kanamycin (1957)b | 2–4 | rrs, eis, whiB7 | 16S rRNA, aminoglycoside acetyltransferase, transcriptional regulator | Inhibition of protein synthesis | 76 |
| Capreomycin (1960)b | 2–4 | rrs, tlyA | 16S rRNA, 20-O-methyltransferase | Inhibition of protein synthesis | 85 |
| Quinolones (1963)b | 0.5–2.5 | gyrA, gyrB | Subunit A of DNA gyrase, subunit B of DNA gyrase | Inhibition of DNA synthesis | 75–94 |
| Ethionamide (1956)b | 2.5–10 | etaA/ethA ethR inhA | Flavin-containing monooxygenase Transcriptional repressor Enoyl-ACP reductase | Inhibition of mycolic acid synthesis | 37 56 |
| PAS (1946)b | 1–8 | thyA dfrA folC ribD | Thymidylate synthetase Dihydrofolate reductase Dihydrofolate synthetase Enzyme in riboflavin biosynthesis | Inhibition of folic acid and thymine nucleotide metabolism | 37 |
| Cycloserine (1955)b | 10–40 | alr | Alanine racemase | Inhibition of cell wall peptidoglycan synthesis | Unknown |
| ddl | d-Alanine–d-alanine ligase | ||||
| cycA | Probably transporter of D-serine/alanine/glycine |
ACP: acyl-carrier-protein; MIC: minimum inhibitory concentration; PAS: para-aminosalicylic acid.
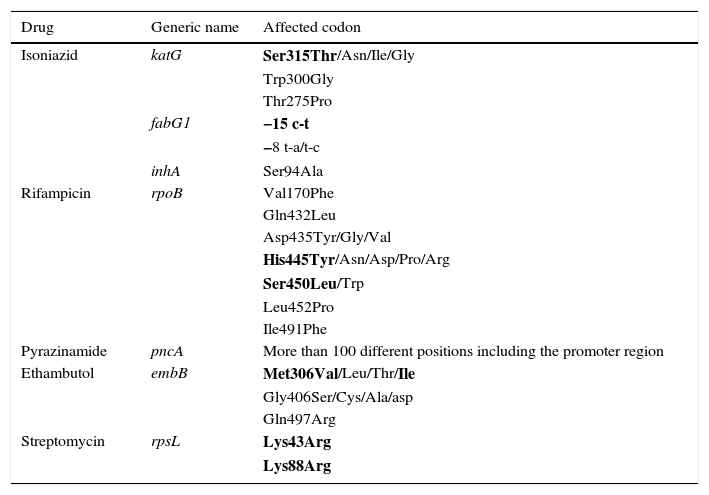
Rifampicin is a major bactericide that, in addition to interfering in RNA synthesis by binding to the β subunit of RNA polymerase, has been reported to induce the formation of hydroxyl radicals in susceptible strains, contributing to an increase in its bactericidal power.28 More than 96% of Rif resistance is caused by mutations in a 81-pb region of the rpoB gene (RRDR codons 428–456 or 507–533 referring to H37Rv or Escherichia coli, respectively) that codifies the β subunit of the RNA polymerase.29 These mutations in the essential protein RpoB would cause a reduction in fitness and would be associated with a lesser transmissibility of the bacteria from one host to another. However, it is suggested that a Rif-resistant strain can have success in transmission, and a high minimum inhibitory concentration (MIC) if it is able to generate efficient compensatory mutations that would allow it to elude or bypass this cost, which would explain why some MDR strains transmit better than others.30 Casali et al. found that 97% of Beijing strains that caused MDR-TB cases in the Russian population had the most common mutation in rpoB, S450L (S531L referring to E. coli), and had compensatory mutations in the rpoA or rpoC genes or in the rpoB gene itself.31 Meftahi et al. also describe this for the MDR Haarlem strain responsible for an outbreak in Tunisia, which had a second mutation in codon V534M confirming with phenotypic testing that this mutation compensated for the fitness lost in addition to increasing the level of resistance to Rif.32 Mutations in rpoB are generally associated with resistance to all rifamycins, including rifabutin, however the 514, D516V and S522L mutations have been described as associated with resistance to Rif and susceptibility to rifabutin. Also, mutations have been found in susceptible strains, for example 510H, L511P, D516Y, N518D, H526N and L533P,33,34 which could cause errors in the molecular techniques if not taken into consideration. Monoresistance to Rif is rare; generally it is associated with resistance to INH. This fact, along with the high performance of its molecular detection, leads to this being used as a good marker for MDR-TB.1Table 2 shows the most frequent mutations consistent with resistance to first-line drugs.
Resistance to first-line drugs.
| Drug | Generic name | Affected codon |
|---|---|---|
| Isoniazid | katG | Ser315Thr/Asn/Ile/Gly |
| Trp300Gly | ||
| Thr275Pro | ||
| fabG1 | −15 c-t | |
| −8 t-a/t-c | ||
| inhA | Ser94Ala | |
| Rifampicin | rpoB | Val170Phe |
| Gln432Leu | ||
| Asp435Tyr/Gly/Val | ||
| His445Tyr/Asn/Asp/Pro/Arg | ||
| Ser450Leu/Trp | ||
| Leu452Pro | ||
| Ile491Phe | ||
| Pyrazinamide | pncA | More than 100 different positions including the promoter region |
| Ethambutol | embB | Met306Val/Leu/Thr/Ile |
| Gly406Ser/Cys/Ala/asp | ||
| Gln497Arg | ||
| Streptomycin | rpsL | Lys43Arg |
| Lys88Arg |
Mutations that present with greater frequency associated with resistance to the corresponding drug are indicated in bold.
High-level resistance to isoniazid is mainly due to mutations in the catalase-peroxidase gene, KatG, that catalyses the oxidation of the prodrug to its active form.35 Low-level resistance is associated with mutations in the inhA gene that encodes an NADH-dependent enoyl-ACP reductase, target of the drug, involved in the synthesis of mycolic acids.36 Other genes related to INH resistance have been reported, such as ahpC and mutations that could compensate for the loss of catalase activity.37
Pyrazinamide (PZ) is an important sterilising drug that shortens TB therapy. Nevertheless, PZ's mechanism of action is not well known. Pyrazinoic acid, the active ingredient in PZ, changes the membrane energy and inhibits the membrane transport function in MTB. Preferential PZ activity against bacilli in a non-replicating state is correlated with low membrane potential and the disruption of the membrane potential by pyrazinoic acid and acidic pH.38 Mutations throughout the pncA gene have been described as a cause of resistance to PZ. M. bovis is characterised by a mutation in amino acid 57 of the pncA gene, which also makes it naturally resistant to PZ. The rpsA and panD genes may also be involved in cases of low-level resistance. In fact, Mycobacterium canettii has a panD mutation that seems to be what confers intrinsic resistance to PZ.27
Ethambutol (EMB) interferes with the cell wall's arabinogalactan synthesis. It targets the arabinosyltransferase enzyme and mutations in the embCAB operon encoding it can cause resistance to EMB. The most common mutation arises in codon 306 of embB in up to 68% of cases.39
The aminoglycosides used include SM, AK, KM and CPM. SM inhibits protein synthesis, binding to ribosomal subunit 30S and thus disrupting the reading of messenger RNA. Resistance is caused by mutations in the rpsL gene that encodes for protein 12S and in rrs that encodes for 16S rRNA, which comprises 50% and 20% of SM resistances, respectively. Mutations in gidB have been reported in SM-resistant strains and cross-resistance has been observed with SM and KM in the whiB7 promoter region. Mutations in position 1400 of rrs have been reported to cause resistance to KM and AK. Mutations in the eis promoter region cause low-level resistance to KM but not to AK.27 Mutations in tlyA and in rrs (A1401G and G1484T) are related to resistance to CPM and KM. Multiple mutations can occur in the rrs gene and cause resistance between these drugs.
The mechanism of action of fluoroquinolones is inhibition of topoisomerase II (DNA gyrase), encoded by the gyrA and gyrB genes. Mutations in the gyrA gene, with greater frequency in codons 94, 90 and 88, are associated with resistance. Mutations in gyrB can grant low-level resistance and double mutations in gyrA and gyrB can give a greater level of resistance. An interesting finding is that the concurrence of two T80A and A90G mutations in gyrA make the strain hypersensitive to most quinolones,40 which can make the study of these resistances more complex.41
Cycloserine acts by inhibiting synthesis of the peptidoglycan of the cell wall, although the resistance mechanisms are still not well defined.
Para-aminosalicylic acid has a mechanism of action that is not well known, which could interfere with the biosynthesis of folic acid. Mutations have been reported in the thyA genes in 40% of the cases and also in the folC and dfrA genes.42
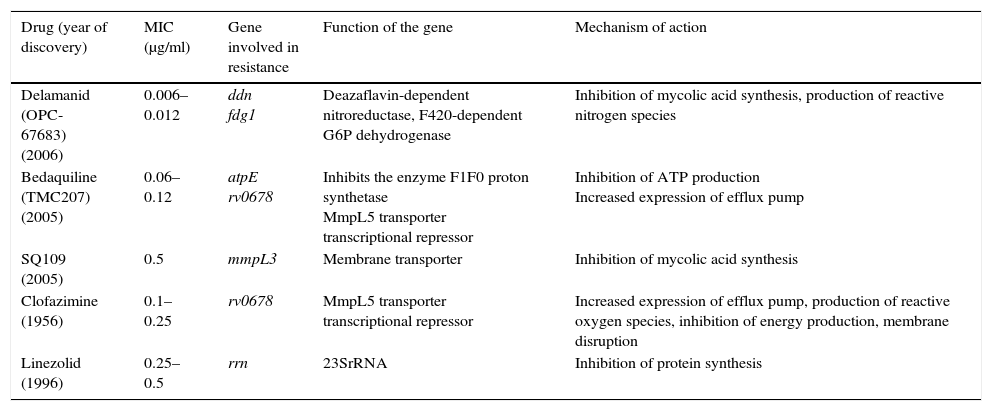
Among the new drugs proposed for the treatment of TB, bedaquiline inhibits adenosine triphosphate (ATP) synthase. It is active against the growing and non-growing population. In the mice models, it seems to have a synergistic effect with PZ. The resistances that have been described would be in atpE and in the Rv0678 gene that would cause overexpression of the MmpL5 efflux pump by causing cross-resistance to clofazimine and bedaquiline. Delamanid inhibits the synthesis of mycolic acids. It must be activated by a nitroreductase enzyme, Ddn, and has activity against bacilli in a non-replicating state. Resistance has been associated with mutations in the fgd1, ddn, fbiA, fbiB and fbiC genes. Oxazolidinones, including linezolid, inhibit protein synthesis binding to subunit 50S of the ribosome and the resistance is mainly associated with mutations in the 23SrRNA gene. SQ109 is an EMB analogue but with a different mechanism of action, which has the protein MmpL3 as its target, making mycolic acid synthesis of the cell wall impossible. Mutations in the mmpL3 gene have been associated with resistance to SQ10927 (Table 3).
Resistance mechanisms of new drugs or proposed agents against Mycobacterium tuberculosis.
| Drug (year of discovery) | MIC (μg/ml) | Gene involved in resistance | Function of the gene | Mechanism of action |
|---|---|---|---|---|
| Delamanid (OPC-67683) (2006) | 0.006–0.012 | ddn fdg1 | Deazaflavin-dependent nitroreductase, F420-dependent G6P dehydrogenase | Inhibition of mycolic acid synthesis, production of reactive nitrogen species |
| Bedaquiline (TMC207) (2005) | 0.06–0.12 | atpE rv0678 | Inhibits the enzyme F1F0 proton synthetase MmpL5 transporter transcriptional repressor | Inhibition of ATP production Increased expression of efflux pump |
| SQ109 (2005) | 0.5 | mmpL3 | Membrane transporter | Inhibition of mycolic acid synthesis |
| Clofazimine (1956) | 0.1–0.25 | rv0678 | MmpL5 transporter transcriptional repressor | Increased expression of efflux pump, production of reactive oxygen species, inhibition of energy production, membrane disruption |
| Linezolid (1996) | 0.25–0.5 | rrn | 23SrRNA | Inhibition of protein synthesis |
ATP: adenosine triphosphate; MIC: minimum inhibitory concentration.
There are various free access databases that collect polymorphisms associated with first- and second-line drug resistance, such as https://tbdreamdb.ki.se/Info/43 and https://umr5558-bibiserv.univ-lyon1.fr/mubii/mubii-select.cgi.
Rapid detection of the genes involved in resistanceThe detection methods are mainly of 2 types, those based on amplification via PCR and solid-phase hybridisation, and those based on real-time PCR.
Solid-phase hybridisation. As we already mentioned in the identification of direct samples, there are 2 tests available, INNO-LIPA1 Rif.TB and Genotype, of which we have 3 versions: MTBDRplus, MTBDRsl and NTM-DR.44 INNO-LIPA has 10 probes on its strip, one which identifies MTBc, 5 which highlight specific regions of the rpoB gene present in the susceptible strains and 4 which detect mutations. It has high sensitivity in cultures and, as always, less in direct samples. The mutations that it can highlight cover approximately 95% of antibiogram confirmed resistances, and serve as a multiresistance marker (MDR-TB)45 since isolated resistance to Rif is quite rare (around 5%). MTBDRplus is similar to the previous test, but also detects resistance to INH by studying the inhA gene (that encodes low-level resistance) and the katG gene (that encodes high-level resistance). As before, it has probes for wild-type regions (susceptible) and their corresponding mutations, covering 80–85% of INH resistances. It has been recommended by the WHO since 2008 and can be performed both on direct samples as well as on cultures, with great sensitivity and specificity.
MTBDRsl, which seeks to detect resistance to second-line drugs such as CP, AK and KM (rss gene), fluoroquinolones (gyrA gene) and EMB (embB gene), with good susceptibility to quinolones, somewhat worse for aminoglycosides and even lower for EMB, have come into existence more recently. There is a second version that includes the study of the eis gene (that improves KM performance, although it worsens the specificity of the injectables), and the gyrB gene for quinolones, with the EMB disappearing.
NTM-DR identifies some NTM species (M. avium, Mycobacterium intracellulare, Mycobacterium quimaera, Mycobacterium chelonae and Mycobacterium abscessus) and detects mutations in the rrs genes for aminoglycosides, and rrl for macrolides, as well as the presence of the erm gene in M. abscessus (inducible resistance to macrolides).
Real-time PCR. As has already been mentioned, real-time PCR has great advantages, therefore it is also entering the area of mycobacteria resistance detection. We have already seen that the Anyplex plus MTB/NTM/DR-TB, in a second step, after a positive result for MTB, can detect resistance mutations. Specifically, it studies 4 mutations for the katG gene and 3 for the inhA gene (resistance to INH), 18 mutations of the rpoB gene (resistance to Rif), 7 mutations of gyrA (quinolones) and 6 mutations for injectable drugs (3 for the rrs gene and 3 for the eis promoter). Its sensitivity and specificity have recently been compared in cultures with MTBDRplus and MTBDRsl being comparable, except for aminoglycosides, which was slightly higher.46 The Abbott RealTime MTB RIF/INH also detects resistance mediated by the same genes (katG and inhA for INH and rpoB for Rif) with very good sensitivity and specificity, having recently received the CE-IVD marking.
Finally, we must mention Xpert MTB/RIF, which, as we have seen, was recommended by the WHO in 2010. It studies the mutations of the rpoB gene, and according to the Cochrane Review,47 it has an accumulated susceptibility of 95% and a specificity of 98%. Its speed and extreme simplicity, since the entire process is integrated into a cartridge that does not require handling, makes it very attractive, having been promoted by the WHO as a tool to rapidly diagnose both the disease as well as the resistant strains in areas with high rates of both. Thanks to a commercial agreement with low-income countries (most of which have these high incidences), it is dispensed at a reduced price, having distributed millions of cartridges. In developed countries, the biggest disadvantage is the price, although in a very recent study it was shown that its use in all patients is cost-effective.48
Whole genome sequencing (WGS) of MTB for the detection of resistance to first- and second-line antituberculosis drugs has already been used on various occasions. A recent review of the topic determined the sensitivity, specificity, positive predictive value, and negative predictive value of WGS using the phenotypic drug susceptibility testing methods as a standard of reference. In conclusion, WGS can be considered a promising alternative to existing phenotypic and molecular susceptibility testing methods for Rif and INH, awaiting the standardisation of the testing procedures.49 A library was also recently published with 1325 polymorphisms based on genomic sequencing that would be predictive of resistance to AM, CP, EMB, ethionamide, INH, KM, moxifloxacin, ofloxacin, PZ, Rif and streptomycin.50
Conflicts of interestThe authors declare that they have no conflicts of interest.
Please cite this article as: Viñuelas-Bayón J, Vitoria MA, Samper S. Diagnóstico rápido de la tuberculosis. Detección de mecanismos de resistencia. Enferm Infecc Microbiol Clin. 2017;35:518–526.