Quinolone resistance in Enterobacteriaceae species has increased over the past few years, and is significantly associated to beta-lactam resistance. The aim of this study was to evaluate the prevalence of chromosomal- and plasmid-mediated quinolone resistance in acquired AmpC β-lactamase and/or carbapenemase-producing Enterobacteriaceae isolates.
MethodsThe presence of chromosomal- and plasmid-mediated quinolone resistance mechanisms [mutations in the quinolone resistance determining region (QRDR) of gyrA and parC and qnr, aac(6′)-Ib-cr and qepA genes] was evaluated in 289 isolates of acquired AmpC β-lactamase- and/or carbapenemase-producing Enterobacteriaceae collected between February and July 2009 in 35 Spanish hospitals.
ResultsPlasmid mediated quinolone resistance (PMQR) genes were detected in 92 isolates (31.8%), qnr genes were detected in 83 isolates (28.7%), and the aac(6′)-Ib-cr gene was detected in 20 isolates (7%). qnrB4 gene was the most prevalent qnr gene detected (20%), associated, in most cases, with DHA-1. Only 14.6% of isolates showed no mutations in gyrA or parC with a ciprofloxacin MIC of 0.5mg/l or higher, whereas PMQR genes were detected in 90% of such isolates.
ConclusionqnrB4 gene was the most prevalent PMQR gene detected, and was significantly associated with acquired AmpC β-lactamase DHA-1. PMQR determinants in association with other chromosomal-mediated quinolone resistance mechanisms, different to mutations in gyrA and parC (increased energy-dependent efflux, altered lipopolysaccharide or porin loss), could lead to ciprofloxacin MIC values that exceed breakpoints established by the main international committees to define clinical antimicrobial susceptibility breakpoints.
En los últimos años se ha observado un incremento de la resistencia a fluoroquinolonas en enterobacterias, estando asociado significativamente a la resistencia a betalactámicos. Nuestro objetivo fue conocer la prevalencia de mecanismos cromosómicos y plasmídicos de resistencia a quinolonas en aislados productores de betalactamasas de clase C adquiridas y/o carbapenemasas.
MétodosSe evaluó la presencia de mecanismos cromosómicos y plasmídicos de resistencia a quinolonas [mutaciones en la región determinante de resistencia a quinolonas de gyrA y parC y genes qnr, aac(6′)-Ib-cr y qepA] en 289 aislados de enterobacterias productoras de betalactamasas de claseC adquiridas y/o carbapenemasas recogidos entre febrero y julio de 2009 en 35 hospitales españoles.
ResultadosSe detectaron determinantes plasmídicos en 92 aislados (31,8%); en 83 aislados (28,7%) se detectó algún gen qnr, y en 20 (7%), la variante aac(6′)-Ib-cr. El gen qnr más prevalente fue qnrB4 (20%), asociado en la mayoría de los casos a DHA-1. El 14,6% de los aislados con una CMI de ciprofloxacino superior a 0,25mg/l no presentaban mutaciones en gyrA ni parC, detectándose en el 90% de los mismos algún determinante plasmídico de resistencia a quinolonas.
ConclusiónqnrB4 fue el determinante plasmídico más prevalente, claramente asociado a DHA-1. Los mecanismos plasmídicos en asociación con mecanismos cromosómicos diferentes a las mutaciones en los genes de las topoisomerasas (sobreexpresión de bombas de expulsión, alteración del lipopolisacárido o disminución de porinas) pueden dar lugar a valores de CMI de ciprofloxacino que superan los puntos de corte establecidos por los principales comités internacionales de definición de puntos de corte para interpretación de datos de sensibilidad.
Fluoroquinolones are broad-spectrum antimicrobials that are used very frequently in clinical practice. For years, high rates of resistance to these antimicrobials have been observed in Enterobacteriaceae worldwide.1 The targets of these compounds are topoisomerase ii (or DNA gyrase), and topoisomerase iv. Resistance to quinolones in enterobacteria may be due to multiple causes that are frequently expressed in the same micro-organism. Traditionally, special attention has been paid to mutations that occur in the gyrA and parC genes encoding subunit A of DNA gyrase and subunit A of topoisomerase iv, respectively; mutations in the gyrB and parE genes, that encode subunits B of the aforementioned enzymes, are less relevant in clinical isolates. In these genes, the mutations associated with quinolone resistance are located in a concrete region thereof called “quinolone resistance-determining region” (QRDR). In gram-negative bacteria, the primary target of quinolones is DNA gyrase, with the mutations first appearing in gyrA and later in parC.1 Despite this, ciprofloxacin-resistant isolates with mutations in parC that are not accompanied by gyrA mutations2 have been reported. A single mutation in gyrA causes resistance to nalidixic acid, but confers a low-level resistance (without exceeding the clinical cut-off point) to ciprofloxacin and levofloxacin.3 By increasing the number of mutations in gyrA and parC, the resistance level also increases until it exceeds the clinical resistance value. Although to a lesser extent, the loss or structural change in the porins, the overexpression of active efflux pumps and changes to lipopolysaccharides also contribute to quinolone resistance.4
To date, 3 types of plasmid-mediated quinolone resistance (PMQR) have been reported: qnr genes, which encode pentapeptide repeat proteins; the variant in one acetyltransferase, AAC(6′)-Ib-cr, which affects piperazinyl group quinolones (for example, ciprofloxacin and norfloxacin), and active OqxAB efflux pump (very common in the Klebsiella pneumoniae chromosome) and QepA.5 In the past 2 decades, a global increase in the prevalence of PMQR has been observed.4
Quinolone resistance is more common in Enterobacteriaceae that are beta-lactam resistant, due to production of extended-spectrum beta-lactamase (ESBL), acquired class C beta-lactamases, or carbapenemases.6 In fact, the first description of a qnr (qnrA1) gene took place in a FOX-5 cephalosporin-producing K. pneumoniae strain.7 Since then, multiple studies have shown the frequent association between genes encoding the aforementioned beta-lactamases, and different variants of genes encoding PMQR, for example, the case of qnrA and qnrB with ESBL from the CTX-M group.4,8
This study analyses the presence of chromosomal mutations (in gyrA and parC) and of PMQRs qnrA, qnrB, qnrC, qnrD, qnrS, qepA and aac(6′)-Ib-cr in acquired class C beta-lactamase-producing and/or carbapenemase-producing Enterobacteriaceae from clinical samples, isolated in a multicentre study with the participation of 35 Spanish hospitals that took place between February and July 2009.9 The objective is to study the association between quinolone resistance mechanisms (chromosomal and plasmid-mediated) with acquired class C beta-lactamases and/or carbapenemases in Enterobacteriaceae, as well as to determine the prevalence of PMQR in this bacterial population.
MethodsBacterial isolatesA collection of 289 isolates from acquired class C beta-lactamase-producing and/or carbapenemase-producing Enterobacteriaceae, including Escherichia coli (n=163), K. pneumoniae (n=57), Proteus mirabilis (n=45), Klebsiella oxytoca (n=10), Enterobacter cloacae (n=9), Citrobacter koseri (n=3), Proteus penneri (n=1) and Salmonella sp. (n=1) was analysed. Only one isolate per patient was included. The clinical and epidemiological data from this collection were previously described.9,10
Antimicrobial susceptibility and study of quinolone resistance mechanismsSusceptibility to nalidixic acid and ciprofloxacin was determined using standardised disc diffusion methods and broth microdilution methods, respectively.11 In nalidixic acid-resistant strains (zone of inhibition diameter ≤13mm)11 or with an MIC for ciprofloxacin of ≥0.25mg/l the mutations in the QRDR of gyrA were analysed using the previously described methodology.12 In strains in which no gyrA mutations were detected, the presence of parC mutations was also studied.12
The presence of qnrA, qnrB, qnrC, qnrD, qnrS, qepA and aac(6′)-Ib genes were analysed in all strains using PCR with the previously-described primers.13 The amplification products were later sequenced.
ResultsPMQR was detected in 92 (31.8%) of the 289 isolates of Enterobacteriaceae analysed using PCR and sequencing. The plasmid-mediated quinolone resistance genes detected included 2 qnrA1, 59 qnrB (58 qnrB4, 1 qnrB19), 10 qnrD, 16 qnrS (8 qnrS1, 8 qnrS2) and 20 aac(6′)-Ib-cr. No qnrC or qepA determinants were detected in any isolate. Seventy-seven isolates (83.7%) presented a single PMQR. The 15 remaining isolates (16.3%) presented two PMQRs: 6 aac(6′)-Ib-cr+qnrS2, 5 aac(6′)-Ib-cr+qnrB4, 3 qnrB4+qnrS1 and 1 qnrB4+qnrS2.
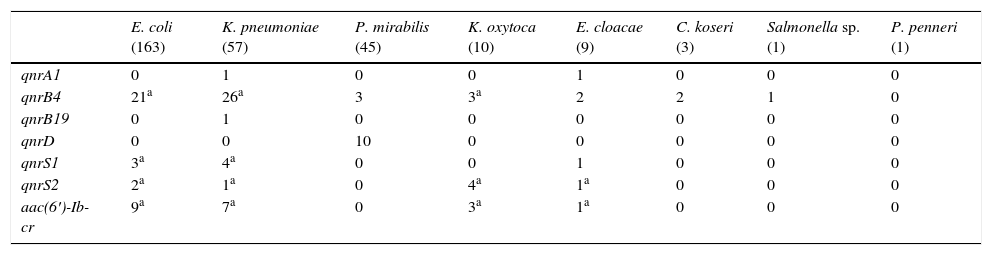
Significant differences were observed in the PMQR type depending on the Enterobacteriaceae species (Table 1). Of the 163 E. coli isolates, at least one plasmid-mediated determinant was detected in 32 isolates (19.6%). The detected determinants were: 21 qnrB4, 5 qnrS and 9 aac(6′)-Ib-cr, carrying 3 two PMQR isolates (2 qnrS2+aac(6′)-Ib-cr and 1 qnrB4+qnrS1). Among the K. pneumoniae isolates, plasmid-mediated mechanisms were detected in 32 (56.1%). A single plasmid-mediated mechanism was detected in 24, and in 8 there were two detected (2 qnrB4+qnrS1, 5 qnrB4+aac(6′)-Ib-cr and 1 qnrS2+aac(6′)-Ib-cr). The PMQR prevalence in the remaining species was: P. mirabilis 28.9%, K. oxytoca 70%, E. cloacae 55.6%, C. koseri 66.7%, Salmonella sp. 100% and P. penneri 0%. The different PMQRs detected in these species are detailed in Table 1. The qnrD gene was only detected in P. mirabilis. The qnrS2 determinant was detected in the presence of another plasmid gene in 7 of the 8 isolates that carried it (87.5%).
Plasmid-mediated quinolone resistance detected by bacterial species.
| E. coli (163) | K. pneumoniae (57) | P. mirabilis (45) | K. oxytoca (10) | E. cloacae (9) | C. koseri (3) | Salmonella sp. (1) | P. penneri (1) | |
|---|---|---|---|---|---|---|---|---|
| qnrA1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| qnrB4 | 21a | 26a | 3 | 3a | 2 | 2 | 1 | 0 |
| qnrB19 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| qnrD | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 |
| qnrS1 | 3a | 4a | 0 | 0 | 1 | 0 | 0 | 0 |
| qnrS2 | 2a | 1a | 0 | 4a | 1a | 0 | 0 | 0 |
| aac(6′)-Ib-cr | 9a | 7a | 0 | 3a | 1a | 0 | 0 | 0 |
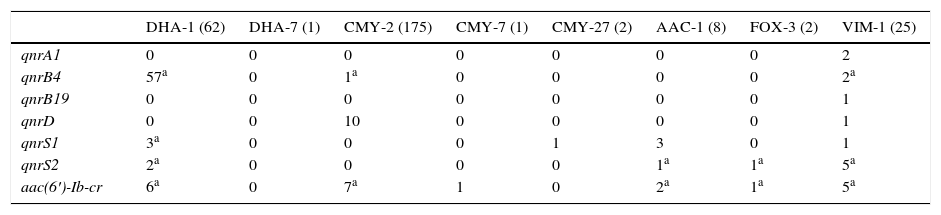
Among carbapenemase-producing isolates (2 IMP-22 producers, one IMP-28 producer and 25 VIM-1 producers) the PMQR prevalence was 39%, detecting some PMQR in 11 isolates of VIM-1 producers: 2 qnrA1, 1 qnrB19, 1 qnrD, 1 qnrS1, 1 qnrS2, 1 qnrB4+aac(6′)-Ib-cr and 4 qnrS2+aac(6′)-Ib-cr.
The prevalence of plasmid-mediated quinolone resistance mechanisms among the 264 acquired class C beta-lactamase-producing isolates was 31.4% (83 isolates). No plasmid-mediated determinants were detected in acquired class C beta-lactamase-producing isolates DHA-6, FOX-8, CMY-4, CMY-48, CMY-55, CMY-57, CMY-59 and CMY-60. The qnrA1 and qnrB19 genes were not detected in acquired class C beta-lactamase-producing isolates. The association between PMQR and acquired class C beta-lactamase is shown in Table 2. The predominant class C beta-lactamase in the collection was CMY-2 (175 isolates), detected in 9.7% of the isolates carrying a PMQR in this enzyme. The 10 qnrD genes were detected in CMY-2-producing P. mirabilis strains. Among the CMY-2-producing Enterobacteriaceae, 7 were detected with aac(6′)-Ib-cr.
Plasmid-mediated quinolone resistance detected based on the acquired class C beta-lactamase and carbapenemase produced.
| DHA-1 (62) | DHA-7 (1) | CMY-2 (175) | CMY-7 (1) | CMY-27 (2) | AAC-1 (8) | FOX-3 (2) | VIM-1 (25) | |
|---|---|---|---|---|---|---|---|---|
| qnrA1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| qnrB4 | 57a | 0 | 1a | 0 | 0 | 0 | 0 | 2a |
| qnrB19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| qnrD | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 1 |
| qnrS1 | 3a | 0 | 0 | 0 | 1 | 3 | 0 | 1 |
| qnrS2 | 2a | 0 | 0 | 0 | 0 | 1a | 1a | 5a |
| aac(6′)-Ib-cr | 6a | 0 | 7a | 1 | 0 | 2a | 1a | 5a |
No isolates producing acquired class C beta-lactamases DHA-6, FOX-8, CMY-4, CMY-48, CMY-55, CMY-57, CMY-59 and CMY-60 and carbapenemases IMP-22 and IMP-28 are included because PMQR was not detected in any of them.
Among the 62 DHA-1-producing isolates, PMQR was detected in 58 (93.5%), with qnrB4 being the most frequently found, present in 57 of said isolates (in 9 isolates, a second plasmid-mediated determinant was also detected). The association between qnrB4 and DHA-1 was statistically significant (p<0.05).
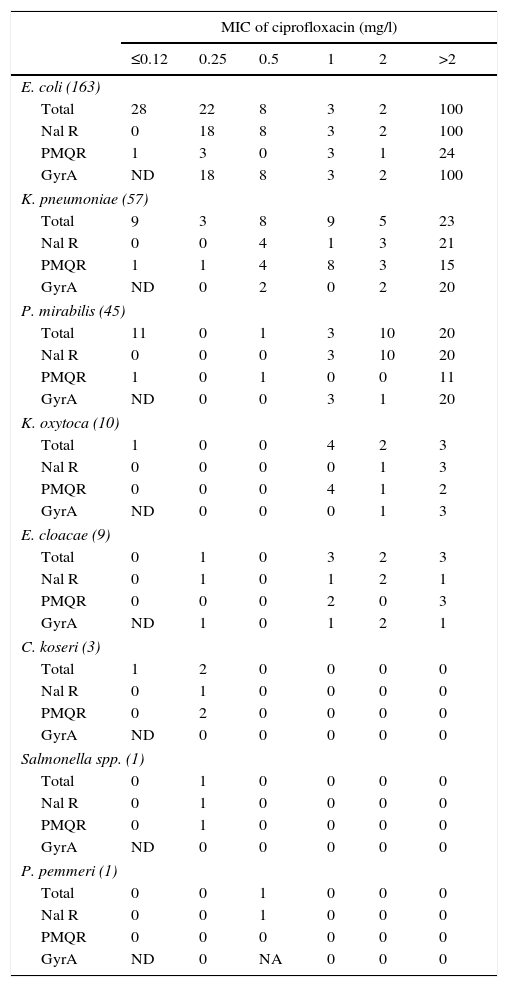
With respect to the chromosomal mechanisms of quinolone resistance, the QRDR of gyrA of the nalidixic acid-resistant isolates,11 or that had an MIC for ciprofloxacin equal to or greater than 0.25mg/l, was analysed. No isolate was found with mutations in parC s without also having it in gyrA. In E. coli., it was verified that all nalidixic acid-resistant isolates showed mutations in the analysed region (Table 3). In this bacterial species there were 4 isolates with MIC for ciprofloxacin of 0.25mg/l that did not present mutations in gyrA or in parC; 3 of them presented PMQR. Also, in this Enterobacteriaceae, isolates were found with mutations in gyrA and PMQR independent of their ciprofloxacin MIC value, something not observed in the isolates from other species, in which only the combination of PMQR and gyrA mutations in isolates with ciprofloxacin MICs exceeding 2mg/l were detected.
Characteristics of the isolates in the collection by ciprofloxacin MIC.
| MIC of ciprofloxacin (mg/l) | ||||||
|---|---|---|---|---|---|---|
| ≤0.12 | 0.25 | 0.5 | 1 | 2 | >2 | |
| E. coli (163) | ||||||
| Total | 28 | 22 | 8 | 3 | 2 | 100 |
| Nal R | 0 | 18 | 8 | 3 | 2 | 100 |
| PMQR | 1 | 3 | 0 | 3 | 1 | 24 |
| GyrA | ND | 18 | 8 | 3 | 2 | 100 |
| K. pneumoniae (57) | ||||||
| Total | 9 | 3 | 8 | 9 | 5 | 23 |
| Nal R | 0 | 0 | 4 | 1 | 3 | 21 |
| PMQR | 1 | 1 | 4 | 8 | 3 | 15 |
| GyrA | ND | 0 | 2 | 0 | 2 | 20 |
| P. mirabilis (45) | ||||||
| Total | 11 | 0 | 1 | 3 | 10 | 20 |
| Nal R | 0 | 0 | 0 | 3 | 10 | 20 |
| PMQR | 1 | 0 | 1 | 0 | 0 | 11 |
| GyrA | ND | 0 | 0 | 3 | 1 | 20 |
| K. oxytoca (10) | ||||||
| Total | 1 | 0 | 0 | 4 | 2 | 3 |
| Nal R | 0 | 0 | 0 | 0 | 1 | 3 |
| PMQR | 0 | 0 | 0 | 4 | 1 | 2 |
| GyrA | ND | 0 | 0 | 0 | 1 | 3 |
| E. cloacae (9) | ||||||
| Total | 0 | 1 | 0 | 3 | 2 | 3 |
| Nal R | 0 | 1 | 0 | 1 | 2 | 1 |
| PMQR | 0 | 0 | 0 | 2 | 0 | 3 |
| GyrA | ND | 1 | 0 | 1 | 2 | 1 |
| C. koseri (3) | ||||||
| Total | 1 | 2 | 0 | 0 | 0 | 0 |
| Nal R | 0 | 1 | 0 | 0 | 0 | 0 |
| PMQR | 0 | 2 | 0 | 0 | 0 | 0 |
| GyrA | ND | 0 | 0 | 0 | 0 | 0 |
| Salmonella spp. (1) | ||||||
| Total | 0 | 1 | 0 | 0 | 0 | 0 |
| Nal R | 0 | 1 | 0 | 0 | 0 | 0 |
| PMQR | 0 | 1 | 0 | 0 | 0 | 0 |
| GyrA | ND | 0 | 0 | 0 | 0 | 0 |
| P. pemmeri (1) | ||||||
| Total | 0 | 0 | 1 | 0 | 0 | 0 |
| Nal R | 0 | 0 | 1 | 0 | 0 | 0 |
| PMQR | 0 | 0 | 0 | 0 | 0 | 0 |
| GyrA | ND | 0 | NA | 0 | 0 | 0 |
GyrA, number of isolates with mutations in the QRDR of gyrA. No isolate was found without mutations in gyrA that had any mutation in parC; NA, the QRDR of gyrA was not amplified; Nal R, number of isolates resistant to nalidixic acid (halo diameter ≤13mm); ND, the presence of mutations in gyrA in the isolates susceptible to nalidixic acid was not determined (inhibition halo diameter >13mm)11 or with a ciprofloxacin MIC≤0.12mg/l; PMQR, number of isolates of plasmid-mediated quinolone resistance carriers; Total, total number of isolates of each species.
Among the isolates of species other than E. coli, 9.5% were found to be resistant to nalidixic acid and did not have mutations in the QRDR of gyrA or parC (Table 3); 66.7% of isolates with a ciprofloxacin MIC equal to or greater than 0.25mg/l had a mutation in gyrA and 52.8% carried some PMQR.
DiscussionThe development and expansion of the beta-lactam resistance, linked to the broad-spectrum of fluoroquinolone antimicrobials, led to the use of these drugs as empirical therapy for a wide variety of infections of community and nosocomial origin. The widespread use of quinolones has caused an increase in fluoroquinolone resistance levels worldwide. In Spain, the rate of fluoroquinolone-resistant E. coli isolates has increased more than 16% in recent years, going from 17.3% in 2001 to 34% in 2014.14 The situation of K. pneumoniae is similar, with the increased levels of resistance being somewhat lower. In this study, 59% (170) of the acquired class C carbapenemase- or beta-lactamase-carrying isolates analysed were not susceptible to ciprofloxacin. This is a concerning piece of information that highlights the reduced number of therapeutic options for multidrug-resistant Enterobacteriaceae isolates.
Fluoroquinolone resistance is mainly due to mutations in the genes of type ii topoisomerases and is a sequential process, such that the appearance of the first mutation in gyrA favours the appearance of new mutations in parC and gyrA that entails an increase in the ciprofloxacin MIC above 2mg/l.15 The rest of the known resistance mechanisms, both chromosomal and plasmid-mediated mechanisms, confer a low level of resistance that does not exceed the cut-off points established by international agencies.5 In the collection analysed in this study, 24 isolates were found (15 K. pneumoniae, 5 K. oxytoca and 4 E. cloacae) with a ciprofloxacin MIC equal to or exceeding 1mg/l that did not have mutations in the QRDR of either gyrA or parC; 23 of these isolates present at least one PMQR. The qnr genes present in these isolates were qnrB4 and/or qnrS (some isolates also carried the aac(6′)-Ib-cr) gene, which are associated with a greater increase in the fluoroquinolone MIC.3,16 According to our data, the presence of isolates with an elevated fluoroquinolone MIC in the absence of gyrA and parC mutations and PMQR carriers has recently been reported in Poland.2
The presence of qnr genes favours the appearance of chromosomal mutations that reduce the permeability of fluoroquinolones or increase their expulsion from the cell interior (by means of reducing the number of porins, changes in lipopolysaccharides or the increase in the expression of efflux pumps), that determines an increase in the ciprofloxacin MIC.17,18 Also, the mutations affecting the entry and exit of quinolones appear before the mutations in the QRDRs,19 something that is consistent with the data obtained in this study, where it is observed that the majority of isolates with a ciprofloxacin MIC between 0.5 and 2mg/l does not result in the combination of PMQR and chromosomal mutations in the QRDRs. On the other hand, the combination of both mechanisms is observed in strains with MICs above 2mg/l. Therefore, the high MIC in these isolates could be due to a synergistic combination of PMQRs and chromosomal mutations reducing the intracellular concentration of quinolones, so that in later steps these isolates could acquire mutations in the QRDR of the gyrA and parC genes, increasing the level of resistance to fluoroquinolones.
One of the current main limitations in the study of the prevalence of PMQRs is the absence of their phenotypic markers,13 which leads to the majority of the prevalence information for these resistance genes being conducted in specific populations: fluoroquinolone-resistant isolates or ESBL producers.20–22 This makes it difficult to compare studies.
In this study, the prevalence of PMQR in acquired class C beta-lactamase- and/or carbapenemase-producing strains was 32%, a similar value to those found in similar collections.23 The data regarding carbapenemase-producing strains indicate a PMQR prevalence of 39%, a value less than those found in other regions of the world, such as China, in carbapenem-resistant strains.24 The information obtained from the carbapenemase-producing isolates must be taken with caution due to the low number of beta-lactamase-producing strains included in this study (28). Also, since 2010, an expansion of carbapenemases has taken place in Spain, mainly due to the dispersion of OXA-48, with this enzyme becoming the most prevalent in this country.25 It would therefore be interesting to know about the prevalence of PMQR in beta-lactamase-producing isolates in Spain. In this regard, the coexistence of OXA-48 and AAC(6′)-Ib-cr in K. pneumoniae has recently been reported in Turkey.26
In our collection, the most prevalent PMQR was qnrB4 (20%), followed by the aac(6′)-Ib-cr gene (7%). This data differs from that which is published in many studies where the most prevalent PMQR is aac(6′)-Ib-cr, followed by qnrB.21,24,27 This discordance is due to the nature of the collection that is the object of this study, where the high prevalence of qnrB4 is due to its association with DHA-1,28,29 having detected qnrB4 in 92% of the class C plasmid-mediated beta-lactamase-producing isolates. A previous analysis in 22 of the strains that were considered for this study had already allowed for verification that in the vast majority of qnrB4 cases the genetic environment of the blaDHA-1 gene was present.30 Given the different populations analysed in the different studies about the prevalence of PMQR conducted worldwide, it is difficult to compare them. It would be interesting to conduct prevalence studies using unbiased bacterial populations, so that we could obtain prevalence data without over- or underestimates. In this respect, an algorithm was published recently that allowed for phenotypic detection of PMQR in strains lacking QRDR modifications,31 which would facilitate the conduct of a study of the prevalence of these resistance mechanisms.
The results obtained support that which is stated by other authors,3,16,32 that although the PMQRs to quinolones alone could only cause a moderate level of resistance to quinolones, their association with chromosomal mechanisms could bring the quinolone MIC values above the established cut-off points. Therefore, it would be beneficial to be familiar with the epidemiology of these plasmid-mediating mechanisms and to be able to control their dispersion, given that they could help reduce or prevent increased resistance to quinolones.
FundingThis study was partially funded by the Ministerio de Sanidad, Servicios Sociales e Igualdad [Ministry of Health, Social Services and Equality], Instituto de Salud Carlos III-FEDER [Carlos III-FEDER Health Institute], Red Española de Investigación en Patología Infecciosa [Spanish Network of Infectious Disease Research] (REIPI RD06/0008), via subsidies from the Fondo de Investigación Sanitaria [Health Research Fund] (PS09/00125 and PI11/01117) and assistance from AstraZeneca Farmacéutica España and Wyeth (now Pfizer).
Conflicts of interestThe authors declare that they have no conflicts of interest.
To the members of the SEIMC [Spanish Society of Infectious Diseases and Clinical Microbiology] GEMARA and GEIH groups.
Please cite this article as: Machuca J, Agüero J, Miró E, Conejo MC, Oteo J, Bou G, et al. Prevalencia en España de mecanismos de resistencia a quinolonas en enterobacterias productoras de betalactamasas de clase C adquiridas y/o carbapenemasas. Enferm Infecc Microbiol Clin. 2017;35:485–490.