In this review we try to update the new procedures applicable in the microbiological diagnosis of bacteriemia and fungemias.
MethodReview of scientific literature.
Results and conclusionsAfter defining the process and indicating its fundamental principles, the main biomarkers used in clinical practice are reviewed. Subsequently, the particularities of the pre-analytical phase (collection and transport of samples) are highlighted and the steps to follow for the microbiological identification by classical methods are detailed, based on the culture of the blood samples. In the following section, we review the diagnostic methods not culture based, including those that detect the presence of the genome of the microorganism and those based on the study of proteome by mass spectrometry. The last section describes the procedures more frequently used for the study of antibiotic susceptibility, both by phenotypic and genotypic methods.
En esta revisión se pretende actualizar los nuevos procedimientos aplicables en el diagnóstico microbiológico de las bacteriemias y fungemias.
MétodoRevisión de la literatura científica.
Resultados y conclusionesTras definir el proceso e indicar sus principios fundamentales, se revisan los principales biomarcadores utilizados en la práctica clínica. Posteriormente, se resaltan las particularidades de la fase preanalítica (recogida y transporte de las muestras) y se detallan los pasos a seguir para la identificación microbiológica por métodos clásicos, basados en el cultivo de las muestras de sangre. En el siguiente apartado, se revisan los métodos diagnósticos no basados en el cultivo, incluyendo los que detectan la presencia del genoma del microorganismo y los basados en el estudio del proteoma mediante espectrometría de masas. En el último apartado se describen los procedimientos a seguir para el estudio de la sensibilidad antibiótica, tanto por métodos fenotípicos como genotípicos.
The detection of bacteraemia and fungaemia is one of the priorities of the Clinical Microbiology Department, given its diagnostic and prognostic relevance. These infections are associated with increased mortality that can range between 10 and 30% according to the series and the type of patient, the source and the initial management. The severity of this clinical entity requires the rapid administration of empirical antimicrobial treatment, based on clinical data and the local epidemiology of resistance. Despite this, the percentage of inadequate treatments may reach up to 25–30%, with this data being especially key in patients with underlying diseases, severe conditions or in centres with high rates of multidrug-resistant microorganisms.1
Bacteraemia is defined as the presence of bacteria in the blood, which is revealed by isolating bacteria from blood cultures. The source of bacteraemia can be diverse depending on the patient's clinical characteristics. The term fungaemia is used to denote the presence of fungi in the blood, generally yeasts from the genus Candida spp. which, although may stem from similar foci to those that cause bacteraemia, frequently occur as a result of catheter infections.2
Currently, sepsis is defined as life-threatening organ dysfunction in a patient caused by the individual's dysregulated response to infection. A new scale has been designed, known as the quick Sepsis-Related Organ Failure Assessment (qSOFA), which exclusively includes clinical criteria, meaning that it is easy to apply at any care level (not only in hospitals). Septic shock is defined as sepsis symptoms which develop with circulatory and cellular abnormalities, as well as metabolic abnormalities which are severe enough to substantially increase mortality to figures above 40%.
The creation of a specific code for the management of sepsis, code sepsis, stems from the fact that this entity is the main infectious cause of death, affecting 100–150 in every 100,000 inhabitants/year, which accounts for more than 50,000 patients/year in Spain, of whom more than a third may die. Sepsis-related mortality may be combated with correct diagnosis and adequate management within the first few hours of onset. Implementing the package of measures recommended by the Surviving Sepsis Campaign successfully reduces morbidity and mortality to figures of around 25%.3
Microbiological diagnosisIt should be taken into account that the limited number of microorganisms present in the blood during an episode of bacteraemia, which tends to range between 10 colony-forming units (CFU)/ml and 104CFU/ml, may be even lower than 0.1CFU/ml in 20% of cases. This characteristic means that only very sensitive techniques can be used in the rapid diagnosis of this infection. Currently, blood culture continues to be the main diagnostic method for bacteraemia, although its practical value is undermined by the delay in obtaining results and because results are not positive in all patients. Its performance is lower in patients on antibiotic treatment or if the infection is caused by fungi, slow-growing bacteria or bacteria with specific requirements. Another key factor is the high proportion of blood cultures contaminated by microorganisms belonging to the skin microbiota. This can result in diagnostic errors, inadequate treatments and lead to high economic costs for the health system. The sensitivity of blood cultures is largely related to the type of microorganism, as well as the volume of the sample, the timing of collection and the absence of previous antibiotic treatments.4
BiomarkersNowadays, we cannot discuss the diagnosis of sepsis without taking into consideration the detection of biomarkers.5 There are some better known biomarkers such as procalcitonin, C-reactive protein, interleukin-6 and mid-regional pro-adrenomedullin, and this is being investigated in a large number of other molecules of diverse origins, including those which reflect changes in the metabolome and some genetic markers of the host (RNA biosignatures) present in serum as a response to bacterial infections. Ideally, these molecules should allow the antimicrobial treatment to be de-escalated or even discontinued in some situations, help establish the severity of the patient's condition and allow their progress to be monitored.6
Processing of blood culturesThere is no universal recommendation as regards the indications for taking blood cultures, although it is generally recommended that they be collected in the presence of chills, fever (body temperature ≥38°C) or hypothermia in neonates and elderly patients. They are also recommended in case of leukocytopenia, leukocytosis or thrombocytopenia not related to haematological processes, in other signs of focal infection or sepsis, as well as in case of suspected endocarditis. Blood culture draws are likewise indicated in small children or elderly patients with a sudden decline in vitality, as these populations may not present with the typical signs and symptoms of bacteraemia. Furthermore, they should always be collected when a catheter tip is sent for seeding due to being suspected as the source of bacteraemia as, if this is not done, it is not possible to differentiate it from catheter colonisation.7
When infection is suspected, blood cultures should be collected from patients who are susceptible to meningitis, osteomyelitis, pyelonephritis, intra-abdominal infection, arthritis, severe skin and soft-tissue infections, pneumonia, endocarditis and fever of unknown origin (occult abscess, typhoid fever, brucellosis, tularaemia, etc.). The blood culture should be accompanied by samples from other sites to try to determine the focus of the infection.
Samples for blood cultures should be collected by venipuncture (peripheral collection) and draws performed with intravascular devices should be avoided, as recommended by the American College of Physicians, with the equipment and anatomical site being changed in each blood culture draw. Draws through a catheter should only be performed if they are intended to diagnose a catheter infection, and this should be accompanied by another draw by peripheral venipuncture. In exceptional circumstances, blood can be collected through a catheter in paediatric patients with haematological cancers. Furthermore, blood should be drawn before antibiotic treatment is initiated and, where this is not possible, when the antibiotic is at its trough concentration (just before the next dose).8
The volume of blood is the most important factor for increasing the diagnostic performance of the blood culture. In general, the recommendations of the Infectious Diseases Society of America/American Society for Microbiology (ASM) are followed, according to which the volume of blood to be cultured is related to the patient's weight. Therefore, in small children, between 1 and 5ml (dilution 1:5) will be required, inoculated in a single aerobic bottle, while the accepted volume of blood to be cultured for older children and adults will be 10–20ml (dilution 1:10), divided between two bottles (anaerobic and aerobic).9
The likelihood of recovering the causative agent increases in relation to the number of blood cultures collected from the patient. It is close to 60–80% in the first blood culture, 80–90% when two blood cultures are collected, and 95–99% in case of a third blood culture. There is no universal recommendation on the interval to be respected between each draw and, although in general it is advised that they be separated by 10–30min, this interval can be shortened in situations of extreme urgency and, in these cases, blood cultures can even be drawn simultaneously from different limbs so as not to delay the antibiotic treatment.
Blood for blood cultures should be drawn under maximal aseptic conditions, scrupulously disinfecting the skin at the site where the venipuncture is going to be performed, as well as the rubber stoppers of the blood culture bottles, preferably with alcoholic 2% chlorhexidine in patients older than two months of age. In all hospitals, the percentage of contaminated blood cultures should be regularly monitored and training activities should be carried out periodically among the staff in charge of blood draws to raise awareness that they should be done in accordance with the protocol established at each centre. Table 1 summarises the method to be followed for collecting blood cultures.
Methodology to be followed for collecting a blood culture.
| 1. Wear gloves and a mask |
| 2. Clean the vial caps with chlorhexidine |
| 3. Select the site from which the blood is to be drawn (do not collect blood through the catheter) |
| 4. Disinfect the skin with chlorhexidine and leave it to work |
| 5. Perform the puncture without touching the patient's skin with your hand |
| 6. Do not allow the needle to come into contact with the cotton wool |
| 7. Draw the blood required so that 10ml of blood can be added to each bottle for adults, and between 1 and 5ml to the paediatric bottle |
| 8. Inoculate the anaerobic bottle first, preventing the entry of air |
| 9. Inoculate the aerobic bottle afterwards; it is not necessary to add air |
| 10. Inoculate the remaining tubes if applicable (biochemistry, etc.) |
| 11. Gently agitate the two bottles |
| 12. Take urgently to the Microbiology Department |
| 13. If this is not possible, keep at room temperature preferably for no more than 2h |
In cases where the bacteraemia source is suspected to be the catheter and it is not easy to replace or do without it, using the differential time to positivity technique is recommended to confirm the diagnosis.10
Once the sample has been obtained and the blood culture bottles have been inoculated, they should be suitably identified in terms of the patient's data and the pairs of bottles corresponding to each of the blood draws. They should be quickly sent to the laboratory and kept at room temperature at all times. The bottles should not be stored in an oven, as it may be that, when they are placed in the smart incubator, they have already reached the stationary growth phase and are thus not detected as positive. It has been reported that there is a significant reduction in pathogen recovery if, instead of placing the bottles immediately in the incubator, they are kept at room temperature over a long period of time. Therefore, the CLSI guidelines recommend that blood culture bottles be placed in the incubator within 2h of when they are drawn. The need to have satellite incubators in places which cannot comply with this requirement should also be assessed.
The development of new automated methods (BacT/Alert® VIRTUO™ [bioMérieux] and BD BACTEC™ FX [Becton Dickinson]) for processing blood cultures has resulted in a substantial breakthrough, as the bottles are placed in automated incubation systems which keep them at a temperature of around 36±1°C. These systems consist in a series of individual cells with continuous agitation to facilitate the multiplication of bacteria, and perform regular monitoring to detect positive bottles.
In addition, each of these companies develops different bottles with particular specifications. Generally speaking, there are bottles designed for the isolation of aerobic bacteria and facultative anaerobes, and bottles for the isolation of facultative and obligate anaerobes. There are also bottles optimised for small volumes of blood, useful in paediatrics, and those selective for mycobacteria (Middlebrook 7H9 broth) or fungi, which can be used in specific cases.
Although the usefulness of the bottles for anaerobes has been questioned due to the low number of obligate anaerobic bacteria responsible for bacteraemia, currently they continue to be recommended in adults as their use helps to locate the focus of the infection and, in addition, 10% of facultative anaerobic bacteria are more quickly recovered in this type of bottle. This is not applicable to the paediatric population, in which the risk of developing bacteraemia due to these microorganisms is much lower.
A total of 85–90% of the blood cultures are positive in less than 48h, except in the event of fungaemia or bacteraemia caused by a slow-growing bacterium. In general, the bottles are incubated for five days before being reported as negative. This time tends to be sufficient for the recovery of most microorganisms, including the fastidious bacteria belonging to the HACEK (Haemophilus spp., Aggregatibacter spp., Cardiobacterium spp., Eikenella spp. and Kingella spp.) group. Incubation should be prolonged in diseases such as endocarditis, which can be caused by slow-growing bacteria, or when the presence of fungi, mycobacteria, Legionella spp., Brucella spp., Bartonella spp. or Nocardia spp. is suspected. It must be taken into account that prolonging the incubation time favours the recovery of contaminants, which is why late findings should be assessed with great caution, always considering the isolated microorganism.11
If the clinical professional and microbiologist must always work together, this is even more the case in light of a suspected infection where the aetiological agent is difficult to isolate and grow. The majority of these bacteria need special culture media due to their nutritional requirements and, in some cases, their incubation should be prolonged because of their slow growth. In some of these infectious processes, serological testing and molecular microbiology techniques are essential procedures for diagnosis.
Yeasts grow well in routine blood culture bottles. However, this test is only positive in approximately half of patients in whom fungaemia is suspected due to the fact that monocytes and other cells of the immune system lyse the fungal cells, there may be a very low inoculum or fungaemia may be transitory or intermittent. In order to improve the technique's sensitivity, the volume of blood cultured is a crucial factor. The usefulness of performing an additional draw for a fungi-specific bottle is disputed, although some authors have reported that they improve recovery rates, particularly for infections associated with Candida glabrata.
In relation to the incubation time of the blood culture bottles with suspected fungal infections due to yeasts, the vast majority of these microorganisms are isolated within the first three days of incubation, with C. glabrata and Cryptococcus neoformans being the pathogens which exhibit the slowest growth. Despite this, due to the low profitability of this diagnostic method in this process, it is advised to extend the incubation time of these bottles if after five days they remain negative and there is a high clinical suspicion of fungaemia.12
A key issue when establishing the clinical significance of microbiological findings is to successfully differentiate between contamination and infection. The United States National Healthcare Safety Network defines a contaminated blood culture (false positive) as one in which species typical of the commensal microbiota of the skin or those typical of the environment are isolated: coagulase-negative staphylococci, other microorganisms of low or zero virulence, such as Aerococcus spp., Micrococcus spp., Propionibacterium acnes, most species of the genera Bacillus and Corynebacterium and some of the viridans group streptococci. However, before considering an isolation of these microorganisms a contaminant, the patient's clinical characteristics and the number of blood cultures in which they are isolated should be carefully analysed as, in some cases, these microorganisms may be involved in bacteraemia. In the case of children, this differentiation is particularly difficult because normally only one draw is performed. Measures should thus be taken to prevent the contamination of the bottles and, in case of doubt, a second sample should be taken.13 The detection of Staphylococcus aureus, Streptococcus pneumoniae, Enterobacteriaceae, Pseudomonas aeruginosa and Candida albicans generally translates to true bacteraemia.
Optimal interpretation of the results of positive blood cultures requires knowledge of the patient's clinical situation, their underlying disease, predisposing factors for infection and, if possible, the antimicrobial treatments which have been administered to the patient. This makes it necessary to review patients’ medical histories, particularly nowadays, when they tend to be computerised and easy to access, with close communication with the physician responsible for the patient being highly recommended in order to report the findings at an early stage and assess the process jointly. Fluid communication channels should therefore be established, using new technological developments. It has been repeatedly demonstrated that rapid communication of the results to multidisciplinary teams which apply this information for proper patient management means that the results provided by the Microbiology Department acquire value in the clinical management of these processes and improve their cost-effectiveness. It is therefore important to strengthen these groups in each hospital and to establish rapid and effective information exchange channels, using all available resources.14
Aetiological diagnosis through non-culture based proceduresMost of the available techniques are based on detecting the DNA of the microorganism causing the bacteraemia/fungaemia. The advantage of these techniques, in addition to their speed, is that they are very helpful when dealing with microorganisms which cannot be cultured or are difficult to grow, and when samples are taken after the antimicrobial treatment has been established. In addition, some allow information to be provided on the aetiology of the infection and on the antibiotic susceptibility of the microorganism involved by detecting some resistance determinants. However, they are expensive and require interpretation by experts as they measure DNAaemia rather than bacteraemia and can detect DNA of non-viable bacteria. It must be taken into account that blood has a large quantity of human DNA compared to bacterial DNA and may present normal inhibitory substances (haemoglobin, heparin, etc.) which affect the efficacy of the reaction, producing both invalid results (due to inhibition of the detection technique) and false negative results. In addition, with respect to the sensitivity analysis of molecular techniques, the number of genomic copies of a microorganism present in a sample has to be considered, rather than the determination of the bacterial load in CFU/ml. This concept would also take into consideration the DNA of dead bacteria or those captured by circulating phagocytic cells. According to this option, it is estimated that in an episode of bacteraemia the number of circulating genetic copies would be between 103 and 104 copies of DNA/ml of blood, a value that would be above the limit of detection of most amplification techniques.15
The application of mass spectrometry (matrix-assisted laser desorption/ionisation time-of-flight [MALDI-TOF]) to Clinical Microbiology has resulted in a revolution in the working protocols of centres that possess this technology. This new method can provide aetiological information at an early stage, a few minutes after performing the Gram stain, and therefore enables early adjustment of the antimicrobial therapy, particularly if local epidemiology of antibiotic resistance is available at the centre.16
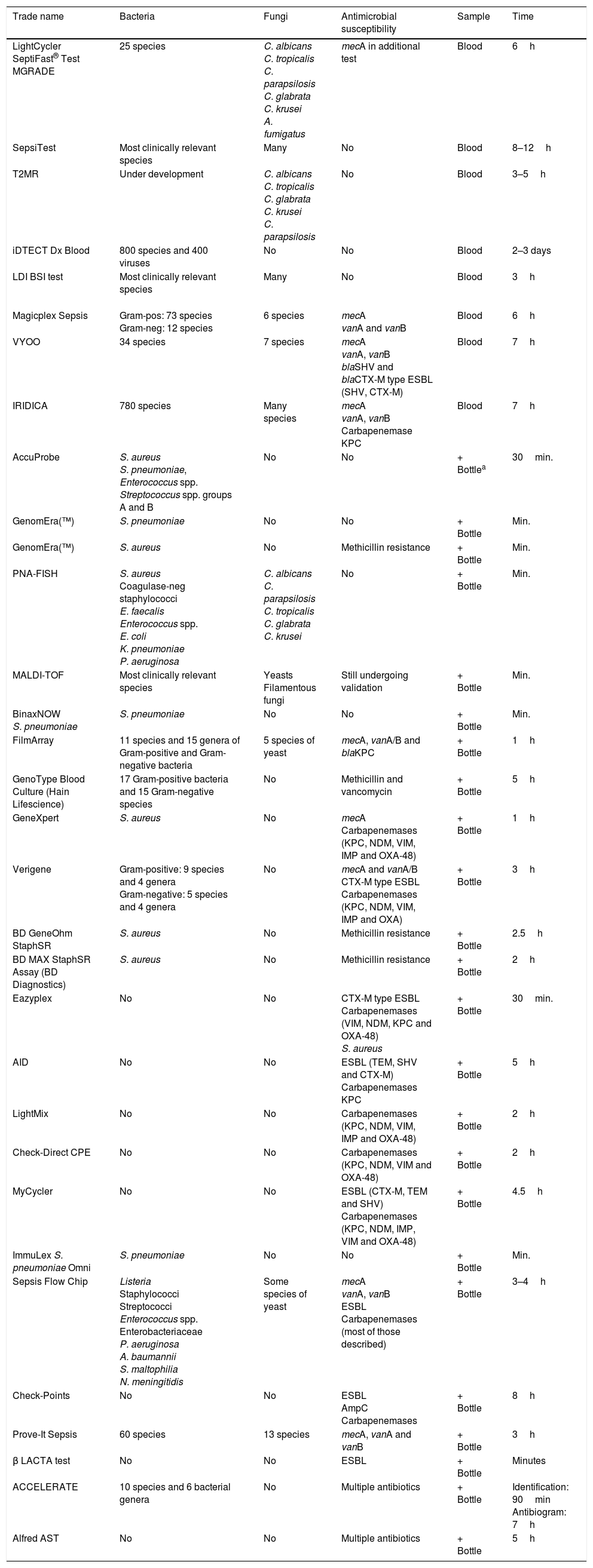
There are two groups of techniques: the first is made up of those applied to bottles of positive blood cultures, in order to obtain information faster than traditional culture-based methods. These techniques are becoming increasingly more widespread as they are of great use in routine clinical practice and correlate closely with the classic methods. A second group of techniques comprises those that are applied directly using the patient's blood, in order to provide information at an even earlier stage. There is less experience with these techniques so they are used to a lesser extent in routine clinical practice17 (Table 2).
Available commercial systems for the detection of bacteraemia and fungaemia.
| Trade name | Bacteria | Fungi | Antimicrobial susceptibility | Sample | Time |
|---|---|---|---|---|---|
| LightCycler SeptiFast® Test MGRADE | 25 species | C. albicans C. tropicalis C. parapsilosis C. glabrata C. krusei A. fumigatus | mecA in additional test | Blood | 6h |
| SepsiTest | Most clinically relevant species | Many | No | Blood | 8–12h |
| T2MR | Under development | C. albicans C. tropicalis C. glabrata C. krusei C. parapsilosis | No | Blood | 3–5h |
| iDTECT Dx Blood | 800 species and 400 viruses | No | No | Blood | 2–3 days |
| LDI BSI test | Most clinically relevant species | Many | No | Blood | 3h |
| Magicplex Sepsis | Gram-pos: 73 species Gram-neg: 12 species | 6 species | mecA vanA and vanB | Blood | 6h |
| VYOO | 34 species | 7 species | mecA vanA, vanB blaSHV and blaCTX-M type ESBL (SHV, CTX-M) | Blood | 7h |
| IRIDICA | 780 species | Many species | mecA vanA, vanB Carbapenemase KPC | Blood | 7h |
| AccuProbe | S. aureus S. pneumoniae, Enterococcus spp. Streptococcus spp. groups A and B | No | No | + Bottlea | 30min. |
| GenomEra(™) | S. pneumoniae | No | No | + Bottle | Min. |
| GenomEra(™) | S. aureus | No | Methicillin resistance | + Bottle | Min. |
| PNA-FISH | S. aureus Coagulase-neg staphylococci E. faecalis Enterococcus spp. E. coli K. pneumoniae P. aeruginosa | C. albicans C. parapsilosis C. tropicalis C. glabrata C. krusei | No | + Bottle | Min. |
| MALDI-TOF | Most clinically relevant species | Yeasts Filamentous fungi | Still undergoing validation | + Bottle | Min. |
| BinaxNOW S. pneumoniae | S. pneumoniae | No | No | + Bottle | Min. |
| FilmArray | 11 species and 15 genera of Gram-positive and Gram-negative bacteria | 5 species of yeast | mecA, vanA/B and blaKPC | + Bottle | 1h |
| GenoType Blood Culture (Hain Lifescience) | 17 Gram-positive bacteria and 15 Gram-negative species | No | Methicillin and vancomycin | + Bottle | 5h |
| GeneXpert | S. aureus | No | mecA Carbapenemases (KPC, NDM, VIM, IMP and OXA-48) | + Bottle | 1h |
| Verigene | Gram-positive: 9 species and 4 genera Gram-negative: 5 species and 4 genera | No | mecA and vanA/B CTX-M type ESBL Carbapenemases (KPC, NDM, VIM, IMP and OXA) | + Bottle | 3h |
| BD GeneOhm StaphSR | S. aureus | No | Methicillin resistance | + Bottle | 2.5h |
| BD MAX StaphSR Assay (BD Diagnostics) | S. aureus | No | Methicillin resistance | + Bottle | 2h |
| Eazyplex | No | No | CTX-M type ESBL Carbapenemases (VIM, NDM, KPC and OXA-48) S. aureus | + Bottle | 30min. |
| AID | No | No | ESBL (TEM, SHV and CTX-M) Carbapenemases KPC | + Bottle | 5h |
| LightMix | No | No | Carbapenemases (KPC, NDM, VIM, IMP and OXA-48) | + Bottle | 2h |
| Check-Direct CPE | No | No | Carbapenemases (KPC, NDM, VIM and OXA-48) | + Bottle | 2h |
| MyCycler | No | No | ESBL (CTX-M, TEM and SHV) Carbapenemases (KPC, NDM, IMP, VIM and OXA-48) | + Bottle | 4.5h |
| ImmuLex S. pneumoniae Omni | S. pneumoniae | No | No | + Bottle | Min. |
| Sepsis Flow Chip | Listeria Staphylococci Streptococci Enterococcus spp. Enterobacteriaceae P. aeruginosa A. baumannii S. maltophilia N. meningitidis | Some species of yeast | mecA vanA, vanB ESBL Carbapenemases (most of those described) | + Bottle | 3–4h |
| Check-Points | No | No | ESBL AmpC Carbapenemases | + Bottle | 8h |
| Prove-It Sepsis | 60 species | 13 species | mecA, vanA and vanB | + Bottle | 3h |
| β LACTA test | No | No | ESBL | + Bottle | Minutes |
| ACCELERATE | 10 species and 6 bacterial genera | No | Multiple antibiotics | + Bottle | Identification: 90min Antibiogram: 7h |
| Alfred AST | No | No | Multiple antibiotics | + Bottle | 5h |
+ Bottle: blood culture bottle with bacterial growth.
Phenotypic studies for the determination of antibiotic susceptibility performed directly on positive blood culture bottles are not endorsed by the EUCAST, CLSI or ASM recommendations, but many studies have reported an excellent correlation between the results obtained from the positive blood culture and those provided by the reference method, which generates the antibiogram using the bacteria colony isolated in solid medium. The use of new automated continuous reading systems in the majority of Microbiology Departments could facilitate the standardisation of the inoculum. With the direct system, the response time is reduced to 24h, with the susceptibility of the bacterium being reported on the same day as the positive blood culture. In any event, considering the preliminary results and subsequently confirming these with standardised methods is recommended.18
Another interesting approach to offer preliminary information is the use of sub-culture plates with chromogenic media for the detection of methicillin-resistant S. aureus, vancomycin-resistant enterococci and extended-spectrum beta-lactamase and carbapenemase-producing Enterobacteriaceae. These media have good sensitivity and specificity, but the results also have to always be confirmed by standardised susceptibility testing methods, so in principle they should only be used when justified by the epidemiological situation.
In relation to non-culture-based methods which provide information on the antimicrobial susceptibility of the microorganism involved in the bacteraemia based on the positive blood culture, new technologies are emerging which must be properly validated before they can be applied to clinical practice. In general, they provide results shortly after the Gram stain has been observed and their data are therefore very useful from a clinical perspective. In order to regulate requests and reduce healthcare costs, it is important for them to be performed in a context of constant communication with the multidisciplinary hospital team responsible for managing bacteraemia/fungaemia at each centre19 (Table 2).
ConclusionsThe rapid and correct diagnosis of bacteraemia/fungaemia should be one of the strategic priorities of all Clinical Microbiology Departments and should form part of a multidisciplinary management system for these diseases, established at each hospital so that all patients can benefit from the results at an early stage. Furthermore, the rapid and correct diagnosis of these processes will contribute to improving antibiotic use, meaning that, as well as reducing healthcare costs, they will help to control antimicrobial resistance.
The necessary measures should be implemented so as much information as possible is available in less than 6h after the blood culture is found to be positive. This information should be passed on urgently to the rest of the multidisciplinary team so that it has a real clinical impact. Indeed, when rapid and efficient communication systems have been applied, they have been shown to have an impact on patients’ progress. In this sense, it is very important that the continuous care model (24h, seven days a week) is advanced in Microbiology Departments, as the impact of an early aetiological diagnosis on patient prognosis is now widely demonstrated.20
Please cite this article as: Guna Serrano MR, Larrosa Escartín N, Marín Arriaza M, Rodríguez Díaz JC. Diagnóstico microbiológico de la bacteriemia y la fungemia: hemocultivos y métodos moleculares. Enferm Infecc Microbiol Clin. 2019;37:335–340.