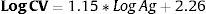Detection of hepatitis C virus (HCV) RNA and the HCV core antigen assay (HCV-Ag) are reliable techniques for the diagnosis of active and chronic HCV infection. Our aim was to evaluate the HCV-Ag assay as an alternative to quantification of HVC RNA.
MethodsA comparison was made of the sensitivity and specificity of an HCV-Ag assay (204 serum samples) with those of a PCR assay, and the correlation between the two techniques was determined.
ResultsThe sensitivity and specificity of HCV-Ag was 76.6% and 100%, respectively. Both assays were extremely well correlated (Pearson coefficient=0.951). The formula (LogCV=1.15*LogAg+2.26) was obtained to calculate the viral load by PCR from HCV-Ag values. HCV-Ag was unable to detect viral loads below 5000IU/mL.
ConclusionAlthough the HCV-Ag assay was less sensitive than the PCR assay, the correlation between both assays was excellent. HCV-Ag can be useful as a first step in the diagnosis of acute or chronic HCV infection and in emergency situations.
La detección del ARN del Virus de la Hepatitis C (VHC) y el ensayo del antígeno del Core del VHC (HCV-Ag) son tèc)cnicas fiables para el diagnóstico de la infección aguda y crónica por el VHC. Nuestro objetivo fue evaluar el HCV-Ag como alternativa a la cuantificación del ARN del VHC.
Mèc)todosAnalizamos la sensibilidad y especificidad del ensayo de HCV-Ag sobre 204 muestras de suero en comparación con un ensayo de PCR, así como su correlación.
ResultadosLa sensibilidad y especificidad del HCV-Ag fueron del 76,6% y 100%, respectivamente. Ambas tèc)cnicas mostraron una excelente correlación (Coeficiente de Pearson=0,951). Obtuvimos una fórmula (LogCV=1,15 * LogAg+2,26) para calcular la carga viral por PCR a partir de los valores de HCV-Ag. El HCV-Ag fue incapaz de detectar viremias por debajo de 5.000UI/mL.
ConclusiónAunque el ensayo de HCV-Ag fue menos sensible que el ensayo de PCR, ambas tèc)cnicas mostraron una excelente correlación. El HCV-Ag puede ser útil como primera etapa en el diagnóstico de la infección aguda o crónica por el VHC y en situaciones de urgencia.
Infection with hepatitis C virus (HCV) is a global health problem that can result in cirrhosis and end-stage liver disease in a substantial proportion of patients.1 Although the initial diagnostic work-up for acute and chronic HCV is usually based on detection of anti-HCV antibodies, anti-HCV assays cannot distinguish between active and past infection. HCV RNA is a reliable marker for the diagnosis of active infection. Moreover, quantification of HCV RNA is useful for monitoring HCV treatment. However, it is costly and requires specialized staff and equipment.
HCV core antigen (HCV-Ag) is an alternative marker found in both complete HCV virions and RNA-free core protein structures. Several HCV-Ag assays have been developed as an affordable alternative to HCV RNA assays for the diagnosis of active HCV infection and for monitoring the therapeutic response. These assays have shown correlations above 90% in most studies.2•7
Our aim was to study a quantitative HCV-Ag assay for evaluation of HCV viremia in order to assess its utility as an alternative to quantification of HVC RNA in routine laboratory practice.
Materials and methodsThe samples used in our study were collected between April 2012 and March 2013. We prospectively performed HCV-Ag testing on serum samples that had been previously characterized by qRT-PCR. These samples were stored in our laboratory at a temperature of ∧80°C. They were not exposed to freezing/de-freezing cycles. We randomly selected samples in order to cover a large variety of viral loads by RT-PCR (range, 18•>108IU/mL). All samples with sufficient serum volume to ensure measurement of HCV-Ag were included. Finally, we selected 204 samples, each corresponding to a single patient.
The HCV genotypes were distributed as follows: 74 samples were genotype 1a; 67 samples were genotype 1b; 41 samples genotype were genotype 3a; 16 samples were genotype 4; and 6 samples were genotype 2. Eight patients out of 204 (3.9%) were undergoing treatment for HCV infection. Given the period when the samples were obtained, no patients were treated with the new direct-acting antiviral agents. All treatment was with interferon (IFN)-based regimens. Five patients (2.6%) were treated with IFN+ribavirin, and the remaining 3 patients (1.3%) were treated with IFN alone. Co-infection with hepatitis B virus (HBV) was recorded in 57 patients (27.9%), HIV infection in 25 patients (12.3%), and co-infection by both HBV and HIV in 19 patients (9.3%).
HCV-Ag was quantified using a fully automated 2-step chemiluminescent microparticle immunoassay (ARCHITECT HCV core-antigen, Abbott Diagnostics, Germany, Wiesbaden). The HCV-Ag assay considers samples as negative below a cutoff of 3fmol/L and positive above 10fmol/L and sets a “gray zone” from 3 to 10fmol/L. Its upper limit is 20,000fmol/L. Samples with a value within the gray zone were considered indeterminate.
HCV-RNA was quantified using the AmpliPre/Cobas TaqMan HCV Test, v 2.0 assay (Roche-Diagnostics, Germany, Mannheim). The linear range for this test extended from 15 to 108IU/mL.
We evaluated the sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of HCV-Ag using HCV-RNA as the gold standard. Results within the quantifiable range of both assays were converted to Log10 values and compared using a Bland•Altman plot. To assess the correlation between HCV-Ag and HCV RNA viral loads, a linear regression analysis was performed by applying the Pearson correlation coefficient and the Deming regression test.8 The statistical analysis was performed using Stata/IC 13.1 (StataCorp, Texas, USA).
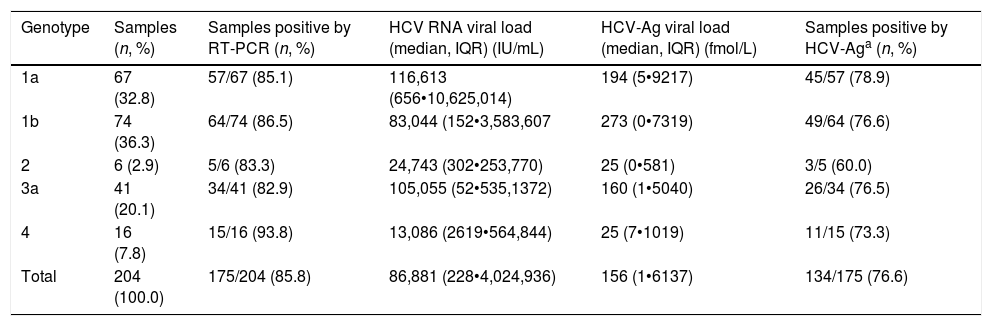
ResultsHCV viremia levels by each technique are summarized in Table 1. HCV RNA was undetectable in 29 samples and detectable in 175 samples. HCV-Ag was performed in all 204 selected samples: overall, 54 samples were negative (<3fmol/L), 16 were indeterminate (3•10fmol/L), and 134 were positive (≥10fmol/L). Of the 29 samples that were negative by RT-PCR, 26 were negative and 3 were indeterminate by HCV-Ag. A total of 41 samples were positive by RT-PCR but negative/indeterminate by HCV-Ag. These 41 samples with HCV-Ag levels below 10fmol/L were shown to have low viral load levels by RT-PCR (median, 201IU/mL; interquartile range, 50•2144IU/mL). HCV-Ag levels were greater than 20,000fmol/L in 23 samples, all of which (100%) had PCR values >107IU/mL (19 samples from 7.06Log to 7.92Log and 4 samples out of the linear range of the PCR assay of >108IU/mL). The sensitivity, specificity, PPV, and NPV of the HCV-Ag assay for predicting detection of HCV RNA were 76.6%, 100%, 100%, and 41.4%, respectively.
HCV viremia levels obtained by both techniques.
| Genotype | Samples (n, %) | Samples positive by RT-PCR (n, %) | HCV RNA viral load (median, IQR) (IU/mL) | HCV-Ag viral load (median, IQR) (fmol/L) | Samples positive by HCV-Aga (n, %) |
|---|---|---|---|---|---|
| 1a | 67 (32.8) | 57/67 (85.1) | 116,613 (656•10,625,014) | 194 (5•9217) | 45/57 (78.9) |
| 1b | 74 (36.3) | 64/74 (86.5) | 83,044 (152•3,583,607 | 273 (0•7319) | 49/64 (76.6) |
| 2 | 6 (2.9) | 5/6 (83.3) | 24,743 (302•253,770) | 25 (0•581) | 3/5 (60.0) |
| 3a | 41 (20.1) | 34/41 (82.9) | 105,055 (52•535,1372) | 160 (1•5040) | 26/34 (76.5) |
| 4 | 16 (7.8) | 15/16 (93.8) | 13,086 (2619•564,844) | 25 (7•1019) | 11/15 (73.3) |
| Total | 204 (100.0) | 175/204 (85.8) | 86,881 (228•4,024,936) | 156 (1•6137) | 134/175 (76.6) |
IQR: interquartile range.
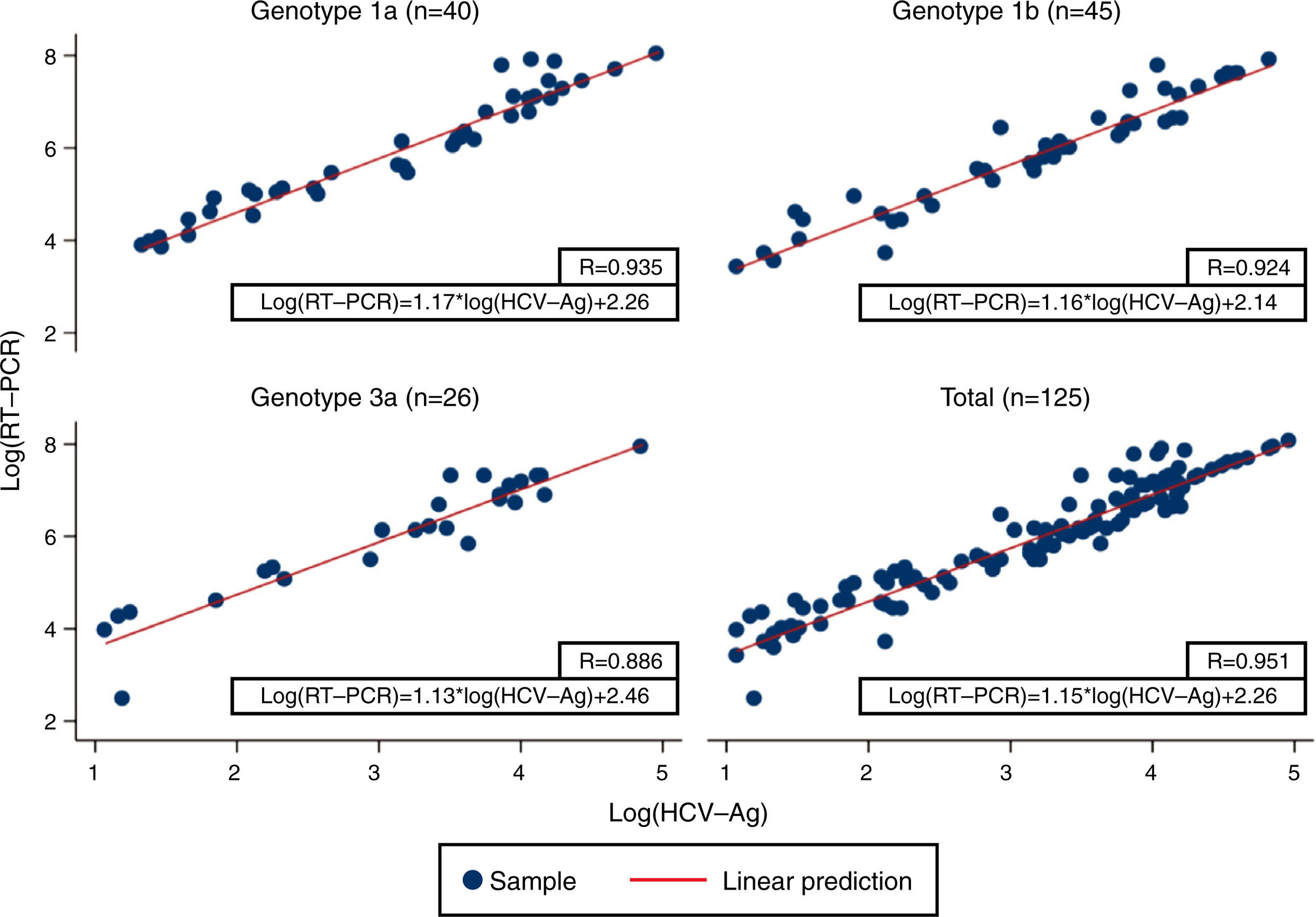
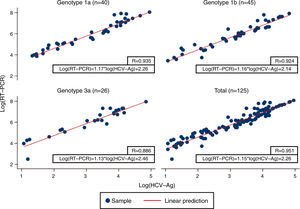
Linear regression analysis was performed in 125 samples, of which 111 corresponded to samples that were within the quantifiable range for both techniques. We also re-analyzed 14 samples that were higher than the upper limit of detection of HCV-Ag. These samples were diluted, and their viral load results were included in the regression analysis. An excellent correlation was observed between the logarithmic values of HCV RNA and HCV-Ag, with a Pearson correlation coefficient of 0.951 (Fig. 1). The expected PCR values from samples were estimated from HCV-Ag values using a simple formula:
where LogCV and LogAg are the logarithmic values for the RNA PCR and HCV-Ag assays respectively.Regression analysis was also performed for each of the most prevalent genotypes (1a, 1b, and 3) (Fig. 1).
DiscussionWe compared the HCV-Ag assay with the reference PCR assay in a group of previously characterized samples. The HCV-Ag assay provided positive results in 76.6% of samples with HCV detected by PCR. In the remaining samples, which tested negative for HCV-Ag, viremia levels (PCR) were generally low. A previous study comparing the ARCHITECT HCV-Ag assay with qRT-PCR9 also reported a lower detection rate for the HCV-Ag assay. A system based on direct detection of viral antigens cannot be compared with another based on amplification of nucleic acids in terms of sensitivity. This drawback is especially important in samples with a low viral load. However, over a specific value, this drawback becomes less relevant, and the correlation between both techniques is high. Although the sensitivity for detecting low HCV RNA viral load in our study was not optimal, the HCV-Ag assay showed excellent specificity and positive predictive value. This finding is in agreement with those of previous reports.5
Previous studies have reported high levels of correlation between HCV RT-PCR and HCV core antigenemia.5,7,10,11 Our findings were in line with those reported in that we obtained a good correlation coefficient (0.951). We also obtained a simple formula that makes it possible to correlate HCV viral load and HCV-Ag based on their logarithmic values. Ottiger et al.,7 obtained similar correlation results in their work. They also found that besides the good correlation between viral load and core antigen data, this correlation was independent of the HCV genotypes. Our findings are comparable, with a high correlation between genotypes 1a, 1b, and 3 (Pearson correlation coefficient: 0.935, 0.924 and 0.886, respectively).
HCV RNA amplification assays are currently the preferred approach for the diagnosis of active HCV infection and for monitoring treatment response. Quantification of HCV-Ag is a less expensive and less complex alternative to the HCV PCR assay. However, it has not been clarified when and how this technique could be useful for the diagnosis and management of HCV infection. In our opinion, the utility of this assay is determined by its main advantages: fast turnaround time, cost, and simplicity. Although widespread use of this marker is not foreseeable, we think it could be useful in specific situations such as screening of organ donors,12 low-income settings, or small and low-skilled laboratories with a limited workload. There are also an increasing number of studies proposing the use of HCV-Ag as a first step in the diagnosis of acute and chronic HCV infection.7,13 Furthermore, recently introduced direct-acting antiviral agents might set the stage to simplify laboratory requirements, and HCV-Ag testing could be used in this subgroup of patients as first-line monitoring viral load or even as a replacement for HCV RNA PCR tests.14 However, more studies are needed to analyze the diagnostic performance of HCV-Ag in the follow up of patients undergoing treatment with direct-acting antiviral agents.
Ethical approvalNo ethical approval was required.
Conflicts of interestThis study does not present any conflicts of interest for the authors.
P. López-Roa has a Río Hortega contract (CM11/00142) from the Instituto de Salud Carlos III-FIS. We thank Thomas O tm)Boyle for his help with the preparation of the manuscript.