Three procedures for rapid identification of microorganisms in positive blood cultures were evaluated.
MethodsWe performed two methods based on direct extraction from a blood culture: Sepsityper® (Bruker Daltonics) (ST) and a non-commercial saponin method (MCS), and another method consisting of a short incubation subculture (SIC). Identification values obtained by spectrometry Matrix-Assisted Laser Desorption Ionization-Time of Flight (EM MALDI-TOF) were compared by applying the manufacturer's interpretation criteria and corrected cut-off points.
ResultsAccording to the manufacturer, 65.8%, 45.8% and 57.4% of microorganisms were identified at the species level by using ST, MCS and SIC, respectively. When applying corrected cut-off points, the values increased to 92.3%, 80.6% and 85.2%, respectively. ST offered significantly better results than MCS, and no significant differences were found between ST and SIC, except for with respect to yeast.
ConclusionsBetter identification rates were obtained by using ST and SIC, which are easily applicable in any laboratory.
Evaluamos tres procedimientos de identificación rápida de microorganismos a partir de hemocultivos positivos.
MétodosAplicamos dos métodos basados en la extracción directa desde el frasco de hemocultivo: Sepsityper® (Bruker Daltonics) (ST) y un método casero con saponina (MCS), y un tercer método basado en un subcultivo con incubación corta (SIC). Se comparan las identificaciones por espectrometría de masas Matrix-Assisted Laser Desorption Ionization-Time of Flight (EM-MALDI-TOF) aplicando los criterios de interpretación del fabricante y los puntos de corte corregidos (PCC).
ResultadosAplicando los criterios del fabricante se identificaron a nivel de especie el 65,8%, el 45,8% y el 57,4% con ST, MCS y SIC, respectivamente. Aplicando los PCC, estos resultados fueron del 92,3%, del 80,6%, y del 85,2%, respectivamente. La identificación con ST fue significativamente mejor que el MCS. ST y SIC no mostraron diferencias significativas, excepto en levaduras.
ConclusionesST y SIC obtienen buenas tasas de identificación y pueden integrarse fácilmente en cualquier laboratorio.
The aim of an effective empirical antibiotic therapy is to improve patient prognosis while reducing length of hospital stay and associated healthcare costs.1
Köck et al.2 found that clinicians changed the empirical treatment in 20% of their patients after identifying the species, versus 8% when only microscopy results were available. In light of these and similar data, new mass spectrometry identification methods like Matrix-Assisted Laser Desorption/Ionisation-Time of Flight (MALDI-TOF) have been developed based on positive blood culture bottles, both direct methods using commercial or in-house extraction techniques,3–10 as well as indirect methods using short-term subcultures.11–14 To interpret the scores obtained by MALDI-TOF, Kohlmann et al.11 published some corrected cut-off points referring to Gram-positive and non-Gram-positive microorganisms.
The primary objective of this study was to evaluate the efficacy of three procedures for the rapid identification of microorganisms in positive blood cultures and their applicability in routine laboratory practice. The second objective was to compare the MALDI-TOF scores by applying the manufacturer's interpretation criteria as well as the corrected cut-off points proposed by Kohlmann et al. to assess their impact on the final identification of the microorganisms.
Material and methodsThis comparative, observational study conducted between 2014 and 2016 evaluated three methods for the rapid identification of pathogens in positive blood cultures.
One bottle per patient and episode of bacteraemia was considered. The BacT/ALERT system (bioMérieux, Marcy l’Etoile, France) was used. The positive bottles were processed following standard protocols. After 24h of incubation, the isolated microorganisms were identified by MALDI-TOF (Bruker Daltonics, Bremen, Germany) and by automated biochemical tests (MicroScan WalkAway, Beckman Coulter, USA). The identity of Streptococcus pneumoniae and viridans streptococci was confirmed by sensitivity to optochin and solubility in bile. Positive blood cultures with Gram stains that revealed polymicrobial aetiology were excluded.
Three aliquots were obtained from the first positive bottle of each patient studied, which were used to evaluate the following procedures:
- 1)
Sepsityper® Kit (ST) (Bruker Daltonics, Bremen, Germany). 1ml of blood culture was processed following the manufacturer's specifications and extraction was performed using ethanol and formic acid. 1μl of supernatant was then placed on a metal plate.
- 2)
In-house saponin method. Based on the protocol modification by Martiny et al.5 1ml was extracted from the blood culture bottle and 500μl of a 5% saponin solution were added. After stirring in a vortex for 10s, it was left to rest for 5min at room temperature. It was washed twice with Milli-Q water. Ethanol–formic acid extraction was then performed and 1μl of the supernatant was placed on a metal plate.
- 3)
Short-term subculture (STS). A chocolate agar medium (Becton Dickinson, Franklin Lakes, NJ, USA) was spiked with 10 drops (≈500μl) of blood culture and incubated in an oven at 37°C, 5% CO2 for 3.5h. After this time had elapsed, a sterile inoculation loop was dragged several times across the surface of the culture and placed on a metal plate. Direct extraction on the plate with formic acid ≥96% was then performed.
1μl of stock solution (α-cyano-4-hydroxycinnamic acid) was added to the inoculum on the plate and left to dry at room temperature. The plate was then inserted in the MALDI-TOF system. The Bacterial Test Standard was used to calibrate the equipment, while the Microflex LT mass spectrometer and the software Biotyper 3.1 (Bruker Daltonics, Bremen, Germany) were used to analyse each sample.
The manufacturer's interpretation criteria defined values ≥2.3 as having a high identification probability, 2–2.2 as probable species identification and 1.7–1.9 as probable genus identification, while values <1.7 were interpreted as unreliable. According to the corrected cut-off points published by Kohlmann et al.,11 correct genus and species identification was assumed with a score ≥1.5 for Gram-positive microorganisms and ≥1.7 for non-Gram-positive microorganisms. As an additional criterion, the first three identification options had to be identical or, failing that, the first two had to be identical with a difference greater than 0.3 with the third. Scores inconsistent with the above were considered to be unreliable. The direct identifications by MALDI-TOF that did not match the definitive identifications of the traditional subculture were considered to be incorrect.
Statistical analysisThe Student's t test with binomial distribution was used to compare the methods used. The total number of correct and unreliable/incorrect identifications were analysed for each method and an analysis by subgroup (Gram-positive microorganisms, Gram-negative microorganisms and yeasts) was also performed.
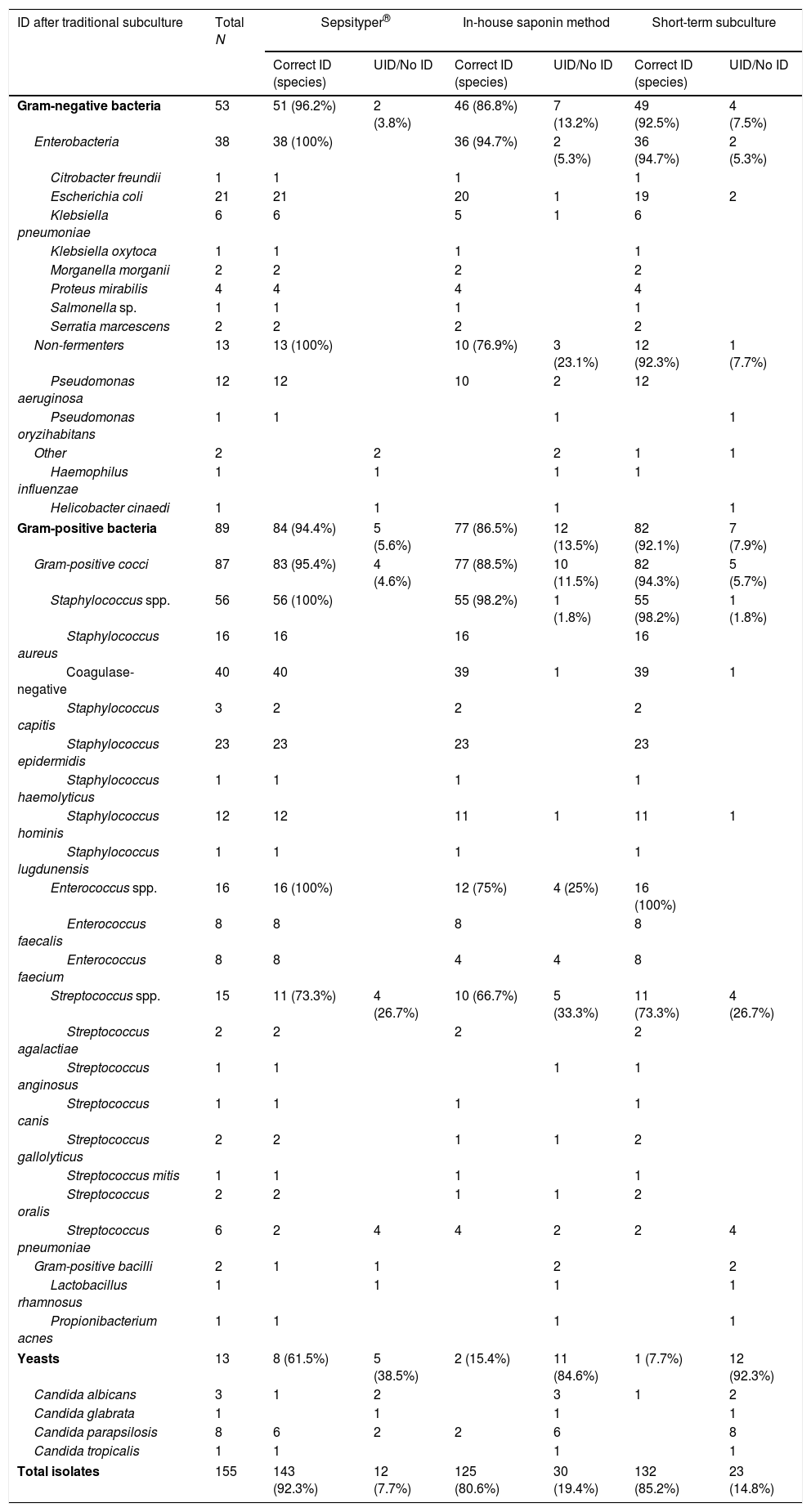
ResultsIn total, 155 blood culture bottles were analysed (90 aerobic and 65 anaerobic): 89 (57.4%) isolated from Gram-positive microorganisms, 53 (34.2%) from Gram-negative microorganisms and 13 (8.4%) from yeasts. Thirty-three (33) different species were identified. The overall results by microorganism and method, applying the corrected cut-off points,11 are shown in Table 1. Table 2 presents the results of each method by interpretation criterion.
MALDI-TOF identification results by microorganism and methods used. The results were interpreted by applying the Kohlmann criteria.
| ID after traditional subculture | Total N | Sepsityper® | In-house saponin method | Short-term subculture | |||
|---|---|---|---|---|---|---|---|
| Correct ID (species) | UID/No ID | Correct ID (species) | UID/No ID | Correct ID (species) | UID/No ID | ||
| Gram-negative bacteria | 53 | 51 (96.2%) | 2 (3.8%) | 46 (86.8%) | 7 (13.2%) | 49 (92.5%) | 4 (7.5%) |
| Enterobacteria | 38 | 38 (100%) | 36 (94.7%) | 2 (5.3%) | 36 (94.7%) | 2 (5.3%) | |
| Citrobacter freundii | 1 | 1 | 1 | 1 | |||
| Escherichia coli | 21 | 21 | 20 | 1 | 19 | 2 | |
| Klebsiella pneumoniae | 6 | 6 | 5 | 1 | 6 | ||
| Klebsiella oxytoca | 1 | 1 | 1 | 1 | |||
| Morganella morganii | 2 | 2 | 2 | 2 | |||
| Proteus mirabilis | 4 | 4 | 4 | 4 | |||
| Salmonella sp. | 1 | 1 | 1 | 1 | |||
| Serratia marcescens | 2 | 2 | 2 | 2 | |||
| Non-fermenters | 13 | 13 (100%) | 10 (76.9%) | 3 (23.1%) | 12 (92.3%) | 1 (7.7%) | |
| Pseudomonas aeruginosa | 12 | 12 | 10 | 2 | 12 | ||
| Pseudomonas oryzihabitans | 1 | 1 | 1 | 1 | |||
| Other | 2 | 2 | 2 | 1 | 1 | ||
| Haemophilus influenzae | 1 | 1 | 1 | 1 | |||
| Helicobacter cinaedi | 1 | 1 | 1 | 1 | |||
| Gram-positive bacteria | 89 | 84 (94.4%) | 5 (5.6%) | 77 (86.5%) | 12 (13.5%) | 82 (92.1%) | 7 (7.9%) |
| Gram-positive cocci | 87 | 83 (95.4%) | 4 (4.6%) | 77 (88.5%) | 10 (11.5%) | 82 (94.3%) | 5 (5.7%) |
| Staphylococcus spp. | 56 | 56 (100%) | 55 (98.2%) | 1 (1.8%) | 55 (98.2%) | 1 (1.8%) | |
| Staphylococcus aureus | 16 | 16 | 16 | 16 | |||
| Coagulase-negative | 40 | 40 | 39 | 1 | 39 | 1 | |
| Staphylococcus capitis | 3 | 2 | 2 | 2 | |||
| Staphylococcus epidermidis | 23 | 23 | 23 | 23 | |||
| Staphylococcus haemolyticus | 1 | 1 | 1 | 1 | |||
| Staphylococcus hominis | 12 | 12 | 11 | 1 | 11 | 1 | |
| Staphylococcus lugdunensis | 1 | 1 | 1 | 1 | |||
| Enterococcus spp. | 16 | 16 (100%) | 12 (75%) | 4 (25%) | 16 (100%) | ||
| Enterococcus faecalis | 8 | 8 | 8 | 8 | |||
| Enterococcus faecium | 8 | 8 | 4 | 4 | 8 | ||
| Streptococcus spp. | 15 | 11 (73.3%) | 4 (26.7%) | 10 (66.7%) | 5 (33.3%) | 11 (73.3%) | 4 (26.7%) |
| Streptococcus agalactiae | 2 | 2 | 2 | 2 | |||
| Streptococcus anginosus | 1 | 1 | 1 | 1 | |||
| Streptococcus canis | 1 | 1 | 1 | 1 | |||
| Streptococcus gallolyticus | 2 | 2 | 1 | 1 | 2 | ||
| Streptococcus mitis | 1 | 1 | 1 | 1 | |||
| Streptococcus oralis | 2 | 2 | 1 | 1 | 2 | ||
| Streptococcus pneumoniae | 6 | 2 | 4 | 4 | 2 | 2 | 4 |
| Gram-positive bacilli | 2 | 1 | 1 | 2 | 2 | ||
| Lactobacillus rhamnosus | 1 | 1 | 1 | 1 | |||
| Propionibacterium acnes | 1 | 1 | 1 | 1 | |||
| Yeasts | 13 | 8 (61.5%) | 5 (38.5%) | 2 (15.4%) | 11 (84.6%) | 1 (7.7%) | 12 (92.3%) |
| Candida albicans | 3 | 1 | 2 | 3 | 1 | 2 | |
| Candida glabrata | 1 | 1 | 1 | 1 | |||
| Candida parapsilosis | 8 | 6 | 2 | 2 | 6 | 8 | |
| Candida tropicalis | 1 | 1 | 1 | 1 | |||
| Total isolates | 155 | 143 (92.3%) | 12 (7.7%) | 125 (80.6%) | 30 (19.4%) | 132 (85.2%) | 23 (14.8%) |
ID: identification; UID/No ID: unreliable identification/no identification.
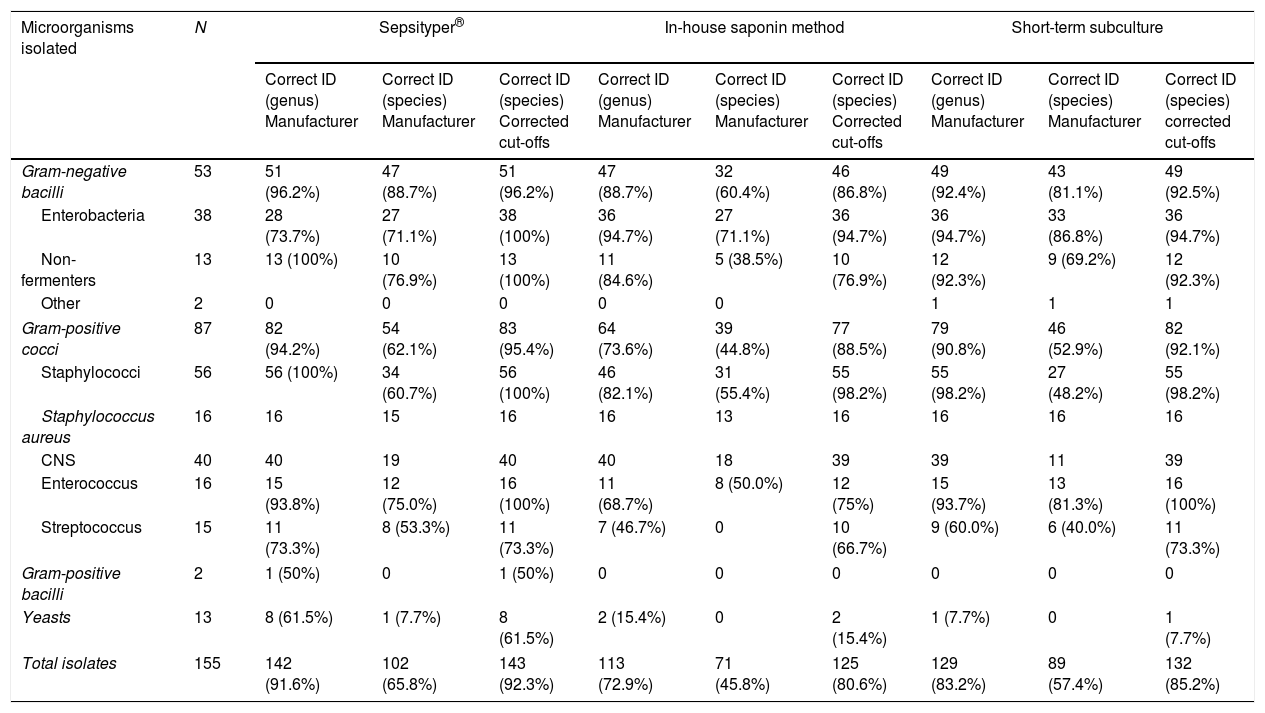
Results obtained by Sepsityper®, the in-house saponin method and the short-term subculture as per the interpretation criteria applied to the scores obtained by MALDI-TOF mass spectrometry.
| Microorganisms isolated | N | Sepsityper® | In-house saponin method | Short-term subculture | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Correct ID (genus) Manufacturer | Correct ID (species) Manufacturer | Correct ID (species) Corrected cut-offs | Correct ID (genus) Manufacturer | Correct ID (species) Manufacturer | Correct ID (species) Corrected cut-offs | Correct ID (genus) Manufacturer | Correct ID (species) Manufacturer | Correct ID (species) corrected cut-offs | ||
| Gram-negative bacilli | 53 | 51 (96.2%) | 47 (88.7%) | 51 (96.2%) | 47 (88.7%) | 32 (60.4%) | 46 (86.8%) | 49 (92.4%) | 43 (81.1%) | 49 (92.5%) |
| Enterobacteria | 38 | 28 (73.7%) | 27 (71.1%) | 38 (100%) | 36 (94.7%) | 27 (71.1%) | 36 (94.7%) | 36 (94.7%) | 33 (86.8%) | 36 (94.7%) |
| Non-fermenters | 13 | 13 (100%) | 10 (76.9%) | 13 (100%) | 11 (84.6%) | 5 (38.5%) | 10 (76.9%) | 12 (92.3%) | 9 (69.2%) | 12 (92.3%) |
| Other | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | |
| Gram-positive cocci | 87 | 82 (94.2%) | 54 (62.1%) | 83 (95.4%) | 64 (73.6%) | 39 (44.8%) | 77 (88.5%) | 79 (90.8%) | 46 (52.9%) | 82 (92.1%) |
| Staphylococci | 56 | 56 (100%) | 34 (60.7%) | 56 (100%) | 46 (82.1%) | 31 (55.4%) | 55 (98.2%) | 55 (98.2%) | 27 (48.2%) | 55 (98.2%) |
| Staphylococcus aureus | 16 | 16 | 15 | 16 | 16 | 13 | 16 | 16 | 16 | 16 |
| CNS | 40 | 40 | 19 | 40 | 40 | 18 | 39 | 39 | 11 | 39 |
| Enterococcus | 16 | 15 (93.8%) | 12 (75.0%) | 16 (100%) | 11 (68.7%) | 8 (50.0%) | 12 (75%) | 15 (93.7%) | 13 (81.3%) | 16 (100%) |
| Streptococcus | 15 | 11 (73.3%) | 8 (53.3%) | 11 (73.3%) | 7 (46.7%) | 0 | 10 (66.7%) | 9 (60.0%) | 6 (40.0%) | 11 (73.3%) |
| Gram-positive bacilli | 2 | 1 (50%) | 0 | 1 (50%) | 0 | 0 | 0 | 0 | 0 | 0 |
| Yeasts | 13 | 8 (61.5%) | 1 (7.7%) | 8 (61.5%) | 2 (15.4%) | 0 | 2 (15.4%) | 1 (7.7%) | 0 | 1 (7.7%) |
| Total isolates | 155 | 142 (91.6%) | 102 (65.8%) | 143 (92.3%) | 113 (72.9%) | 71 (45.8%) | 125 (80.6%) | 129 (83.2%) | 89 (57.4%) | 132 (85.2%) |
CNS: coagulase-negative staphylococci; ID: identification.
With the exception of two strains that were initially identified as viridans group streptococci before being ultimately identified as S. pneumoniae by manual testing, all the identifications obtained by the three study methods matched the identification of the traditional subculture.
The microorganism identification results from the ST were significantly better (p<0.05) than the in-house saponin method. No significant differences were found for either Gram-negative (p=0.094) or Gram-positive microorganisms (p=0.234) in the ST and STS. However, the yeast analysis yielded a p<0.05 in favour of ST. Finally, significant differences (p<0.05) were found in the identification of both Gram-negative and Gram-positive microorganisms after comparing the in-house saponin method with the STS, with the latter producing better results.
DiscussionWe analysed the effectiveness of three methods for the rapid identification of microorganisms in positive blood cultures. When the manufacturer's criteria were applied, the ST (65.8%) and the STS (57.4%) obtained the best species identification results. Significant differences were only found in the yeast identification parameter, with the commercial method yielding the best results. The results of the in-house saponin method were significantly inferior (45.8%) to the other methods.
When the corrected cut-off points11 were applied, the species identification percentages increased: ST (92.3%), STS (85.2%) and in-house saponin method (80.6%), which confirms that the manufacturer's cut-off points can be modified without losing specificity.12,15
The results for the ST are similar to those already published, with correct species identification ranging from 59.5% to 82.8% when the manufacturer's criteria are applied.3–6 Our STS results are similar to those published in the literature.11,13,14 In terms of the in-house saponin method, the protocol amendments proposed by Martiny et al.5 yield better results when the corrected cut-off points11 are applied.
When the corrected cut-off points11 are applied, the Gram-negative bacteria species identification results of both the ST and the STS are excellent (96.2% and 92.5%, respectively). The overall Gram-negative bacteria identification results for the in-house saponin method were statistically inferior (p<0.05), while the Enterobacteria results were comparable with the other two methods.
The ST and STS methods also obtained optimal results for Gram-positive bacteria when the corrected cut-off points11 were applied (94.4% and 92.1%, respectively), with better identification of staphylococci and enterococci than streptococci. Not being able to distinguish between S. pneumoniae and viridans group streptococci is relatively common due to the similarity of their protein spectra. It is noteworthy that all strains of Staphylococcus aureus were correctly identified by the three methods and that no coagulase-negative staphylococcus was erroneously identified as S. aureus.
The yeast identification results were inferior to the rest, with the ST (61.5% applying the corrected cut-off points11) statistically superior to the other methods.
In terms of the applicability of these techniques in laboratory practice, the ST is rapid (20min) but entails a longer technical processing time and higher financial costs. The STS takes longer (3.5h) but requires less technical processing and is cheaper, which means it could easily be integrated into routine laboratory practice.
Therefore, since the identification rates of the ST and STS methods are similar (except for yeasts), both techniques could be used in combination depending on the time to positivity of the blood culture and laboratory logistics, so as to maintain continuous clinical information with sustainable resource use.
Conflicts of interestThe authors declare that they have no conflicts of interest.
Please cite this article as: Martín-Pujol O, Tosco-Nuñez T, de Miguel-Martinez I. Comparación de tres procedimientos para la identificación rápida de microorganismos causantes de bacteriemias. Evaluación de su eficacia y aplicabilidad en el laboratorio de microbiología. Enferm Infecc Microbiol Clin. 2019;37:319–323.