Employing Illumina Hiseq whole genome metagenome sequencing approach, we studied the impact of Trichoderma harzianum on altering the microbial community and its functional dynamics in the rhizhosphere soil of black pepper (Piper nigrum L.). The metagenomic datasets from the rhizosphere with (treatment) and without (control) T. harzianum inoculation were annotated using dual approach, i.e., stand alone and MG-RAST. The probiotic application of T. harzianum in the rhizhosphere soil of black pepper impacted the population dynamics of rhizosphere bacteria, archae, eukaryote as reflected through the selective recruitment of bacteria [Acidobacteriaceae bacterium (p=1.24e−12), Candidatus koribacter versatilis (p=2.66e−10)] and fungi [(Fusarium oxysporum (p=0.013), Talaromyces stipitatus (p=0.219) and Pestalotiopsis fici (p=0.443)] in terms of abundance in population and bacterial chemotaxis (p=0.012), iron metabolism (p=2.97e−5) with the reduction in abundance for pathogenicity islands (p=7.30e−3), phages and prophages (p=7.30e−3) with regard to functional abundance. Interestingly, it was found that the enriched functional metagenomic signatures on phytoremediation such as benzoate transport and degradation (p=2.34e−4), and degradation of heterocyclic aromatic compounds (p=3.59e−13) in the treatment influenced the rhizosphere micro ecosystem favoring growth and health of pepper plant. The population dynamics and functional richness of rhizosphere ecosystem in black pepper influenced by the treatment with T. harzianum provides the ecological importance of T. harzianum in the cultivation of black pepper.
Plants contribute to the establishment of specific ecological niches of microbes in the rhizosphere by playing key role as ecosystem engineers.1 The microbial community at the rhizosphere reflects its functional specificity at the level of plant–microbe interactions. It suggests that taxonomically-contrasted plant growth promoting strains may coexist in soil and colonize the same rhizosphere. The probiotic community enrichment by the plant is the major element in plant response to various biotic and abiotic stresses, coupled with the application of plant growth promoting microbes.2 In the plant rhizosphere, the plant growth-promoting microbes play main roles such as modifying the root functioning, improving plant nutrition and its intake, and influencing the physiology of entire plant. Secondary metabolites secreted by the soil microbes has role in controlling biotic interactions.3 The chemical ecology research field that focus on the understanding the specific interaction mediated by the producer organism with the target microbe and with the microbial community is of immense importance in rhizosphere microniche. The experimental approaches on the role of secondary metabolites suggests that they can act to slow down the germination of spores in order to bring less competitive environment for the growth, act as agents of symbiosis and competitive weapons against other competing organisms.4 Hence, integrating functional and ecological knowledge on microbial populations in soil will be a prerequisite in developing novel management strategies for sustainable agriculture for which the population abundance of soil microbiome is an important component.
Trichoderma (telemorph Hypocrea) is an asexual fungal genus inhabiting the soil of all climatic zones; many of its species are used as effective biofertilizer and biocontrol agents for plants grown in greenhouse as well as fields.5–7 The mechanism mediated by Trichoderma spp. includes the antibiotic activity,8 mycoparasitism,9 cell wall-lytic enzyme action,10 competition for nutrients,11 the induction of systemic resistance to pathogens in plants5; and nutrient supply through the degradation of biomass.6,7
Black pepper (Piper nigrum L.) – a native to India and popularly known as the king of spices – is an export oriented important spice crop grown in tropical countries. The foot rot disease caused by Phytophthora capsici, an oomycete pathogen contributes to the major crop loss as it infects the vine both in nursery and fields.12 The elegant studies on Trichoderma harzianum (MTCC 5179) toward its growth promotion13,14 and disease suppression15–17 activities made this fungus an important component in the integrated disease management module of the cultivation strategy of black pepper in India. Thus, we hypothesized that the probiotic application of Trichoderma would alter the community composition or dynamics of other soil fungi and bacteria at the rhizosphere of black pepper; and that might contribute to the plant health in a better way than the rhizosphere community without Trichoderma. In the light of this hypothesis, this study is designed with three objectives: (a) to inoculate the rhizosphere of black pepper with T. harzianum (MTCC 5179) for assessing its impact on microbial community dynamics in the rhizosphere, (b) to subject the rhizhosphere soil to whole genome metagenomics analysis, and (c) to bring out the taxonomic and functional abundance for understanding the community dynamics.
Materials and methodsRaising of explantSingle node cuttings from Sreekara variety of black pepper were washed with Tween 20 for 15min, followed by running tap water. The cuttings were subsequently surface sterilized with copper oxychloride (0.2%) for 15min, and washed twice with sterile double distilled water (ddH2O). The cuttings were again surface sterilized with mercuric chloride (0.1%) for 5min, followed by wash with ddH2O twice. The cut ends of the cuttings were quick dipped in indole-3-butyric acid (8000ppm), and planted in protray on sterile perlite medium fortified with sterile Hoagland's solution.18
The protrays with the preparation as above were maintained in greenhouse with top portion sealed with aluminum foil. The cuttings were sprayed with Hoagland solution once in a day. After 2 months of growth (when plants attained 24–26cm height with 4–5 leaves), the rhizosphere (perlite) samples from the plants were collected and analyzed for the presence or absence of Trichoderma spp. by plating (spread/pour plate method). Subsequently, saplings with no association of Trichoderma spp. were transferred to the pots filled with top soil (composition: 197 Ca; 173 K, 71 Mg; 18 S; 11.38 Fe; 5.56 Mn; 3.24 Zn; 1.64 P; 0.92 Cu; 0.16 B (all in ppm); and 1.6% organic carbon, pH: 4.35). Two sets of experiments [inoculated with T. harzianum (MTCC 5179), the treatment and without inoculation of T. harzianum, the control] with 4 replicates having 3 plants per replica were designed for the study. Talc formulation of T. harzianum (MTCC 5179) (3.5g/3kg soil) was used for inoculating the soil. Growth parameters viz., height of the plant, stem girth (1cm above from the soil region) and the leaf area index (LAI) were recorded. The LAI was calculated using the formula: length (cm) × width (cm) × 0.6. After 120 days, plants were uprooted, the rhizosphere soil (adhered to the roots of pepper plants) sample were collected from 3 biological replicates of both treatment and control, and stored at −80°C. The weights of shoot and root (fresh and dry) were also recorded.
Extraction of rhizosphere soil DNA and sequencingThe rhizosphere soil DNA from the treated and control plants were extracted from 100mg of soil using MoBio kit (MO BIO Laboratories, Inc. USA), according to the instruction of the manufacturer. DNA from three biological replicates was pooled for the downstream analysis. The integrity of the DNA was assessed by nanodrop spectrophotometer (2000/2000C, Thermo Scientific, USA), and 2μL of each sample was subjected to electrophoresis on 1% agarose gel using 1× tris-borate-EDTA buffer. Gels were stained with ethidium bromide and viewed using Gel imaging System (Syngene Technologies Inc, USA). DNA library was prepared using NEB Next ultra DNA library prep kit for Illumina. Sequencing of the paired end library was done using Illumina Hiseq sequencing platform.
Read quality assessmentThe paired end reads generated were examined for read length, total number of reads, percentage of GC content and mean base quality distribution using FastQC tool kit. All reads were quality filtered with an average Phred quality of 20, and cutadapt (version 1.8.3) was used for adapter removal from the sequences.
De novo assembly and annotationAssembly was performed with default k-mer length (31-size) using de-bruijn graph method. Inhouse PERL and Python code were used to parse the fastq files for the downstream analysis. The sequences were assembled with RayMeta19 using a k-mer size of 31. The contigs with more than 150bp were filtered and taken as pre-processed reads for downstream analysis. Glimmer-MG v 0.3.220 was used to predict the protein coding regions in the contigs. Each sample reads was completely assembled in about 5 days. This run time included de novo contig and scaffold assembly process.
Taxonomy/functional analysisThe taxonomy tree was generated based on neighbor-joining method using MEGAN software. The hierarchy of comparative taxonomic abundance in all the samples was based on the contig abundance with the number of reads assigned to the taxonomy. Functional annotation was performed using DIAMOND version 0.7.921 for predicted genes against the protein database using the BLAST version 2.2.29+22 with an e-value of 1e−5. The functional analyses of all hits were analyzed using the KEGG and SEED options provided in the MEGAN software.23
Analysis by MG-RASTThe results from the standalone workflow were compared with MetaGenome Rapid Annotation using Subsystem Technology (MG-RAST).24 Taxonomic classification was performed to view the taxonomic level in the samples against the M5NR public database using best fit classification with 1e−5 as maximum e-value cutoff, and 60% as minimum identity cutoff. Functional analysis for the distribution of functional categories using subsystems was carried out using the hierarchical classification with 1e−5 as maximum e-value cutoff, and 60% as minimum identity cutoff. Alpha diversity present in the treatment and control samples were estimated.
StatisticsFor the growth parameters, the experimental design adopted was completely randomized design, and the data were analyzed by t-test. Analyses of differential/relative abundance features (of metagenome data) were done using STAMP software package.25 The differential abundance between the samples was calculated using G-test (w/Yates’) + Fisher's test for two sample analysis in STAMP tool.
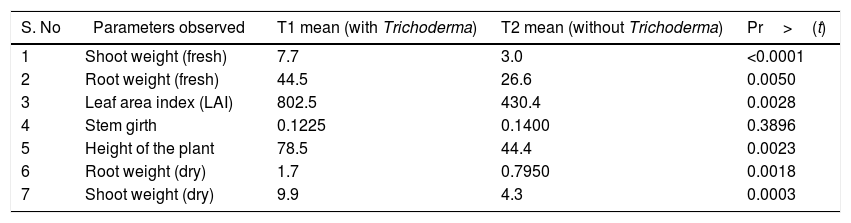
ResultsGrowth parametersThe pH of T. harzianum treated soil was 5.2, after 120 days of inoculation; while that of control was 4.6. Growth parameters, viz., the fresh root, fresh shoot, dry root, dry shoot, LAI (Leaf Area Index), height of the plant were significantly increased in the treatment (Table 1).
Table showing the growth parameters of black pepper: with (treatment) and without (control) inoculation of T. harzianum. The growth parameters at 120 days are shown in the table (n=12).
| S. No | Parameters observed | T1 mean (with Trichoderma) | T2 mean (without Trichoderma) | Pr>(t) |
|---|---|---|---|---|
| 1 | Shoot weight (fresh) | 7.7 | 3.0 | <0.0001 |
| 2 | Root weight (fresh) | 44.5 | 26.6 | 0.0050 |
| 3 | Leaf area index (LAI) | 802.5 | 430.4 | 0.0028 |
| 4 | Stem girth | 0.1225 | 0.1400 | 0.3896 |
| 5 | Height of the plant | 78.5 | 44.4 | 0.0023 |
| 6 | Root weight (dry) | 1.7 | 0.7950 | 0.0018 |
| 7 | Shoot weight (dry) | 9.9 | 4.3 | 0.0003 |
Paired End (251bp × 2) sequencing yielded 2,121,934 and 2,123,836 reads for treatment and control samples, respectively. Majority of the sample reads had 40–70% GC content. The Phred score distribution (≥Q30) of the paired-end metagenome reads for treatment was 79.22%, while 80.82% was for the control. The assembly of reads formed 1,827,461 and 1,879,703 contigs and N50 of 210 and 212, respectively in treatment and control.
Analysis by MG-RASTOut of 4,121,006 (97.1%) sequences that passed quality control, 93.5% sequences produced 3,389,349 predicted protein coding regions of the metagenome in the treatment. Of these, 33.7% sequences were assigned with annotation by M5NR database; 76.0% of annotated features from M5NR database were assigned with functional categories. From control sample, out of 4,162,647 sequences passed quality control (98%), 94.5% produced 3,558,779 predicted as protein coding region. Of these, 33.9% were assigned with annotation by M5NR database, and 74.7% of annotated features were assigned to functional categories. The mean sequence length, mean GC content for treated and control were 248±13bp, 63±7% and 249±12bp, 62±8%, respectively. The double approach we used (stand alone and MG-RAST) for the analysis of metagenome yielded coherent results in both taxonomy and functional categories. The comparative analysis on these metagenomes using MG-RAST is discussed further.
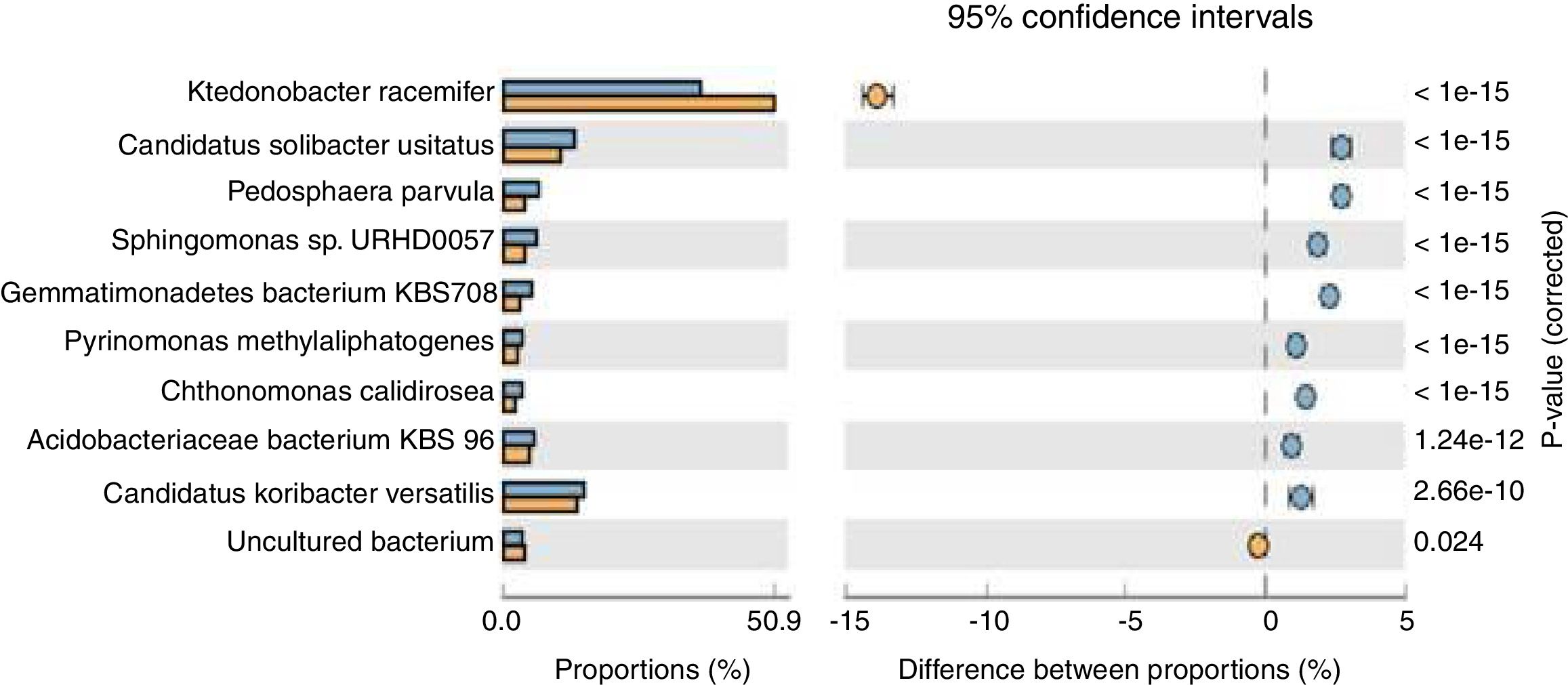
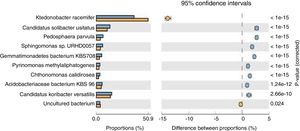
Population dynamicsThe alpha diversity (Shanon diversity index) of the metagenome of both treatment and control samples were 489,569 and 455,862 species, respectively. From the analysis of relative abundance (percentage proportion) for top 10 bacterial species, viz., Acidobacteriaceae bacterium KBS 96, Candidatus koribacter versatilis, Ktedonobacter racemifer, Candidatus solibacter usitatus, Pedosphaera parvula, Sphingomonas sp., URHD0057, Gemmatimonadates bacterium, Pyrinomonas methylalipathogens, Chthonomonas calidirosea and uncultured bacteria [of which A. bacterium (p=1.24e−12) and C. koribacter versatilis (p=2.66e−10) showed statistical significance] were found abundant in the treatment, while uncultured bacteria found were more in control sample (p=0.024) (Fig. 1). The abundance of these bacteria suggests that probiotic application of T. harzianum in black pepper imparted the rhizosphere competence for the bacteria to colonize the roots as the presence of A. bacterium and C. koribacter versatilis has proven as the major rhizosphere competent bacteria involving unique metabolic pathway at the rhizosphere. Analysis of the relative abundance of top 10 fungi, viz., Rhizophagus irregularis, Fusarium oxysporum, Oidiodendran maius, Pseudogymnoasus pannorum, Talaromyces stipitatus, Pestalotiopsis fici, Mortierella verticillata and T. harzianum showed that F. oxysporum (p=0.013), T. stipitatus (p=0.219) and P. fici (p=0.443) were high in treatment, while the control showed higher abundance of R. irregularis (p=0.034), Pseudogymnoasus pannarum (a human pathogenic fungus, p=0.488) and Oidiodendran (p=0.484). The Trichoderma reads were recorded only in treatment sample. The higher abundance of F. oxysporum, T. stipitatus and P. fici in treatment suggests that T. harzianum selectively enriches the biocontrol fungi in the rhizosphere. The reduction of pathogenic fungi, in turn, provides strong evidence that T. harzianum is able to reduce the human pathogenic effect of the amended soil, in comparison to the control.
Species level extended error bar chart profile for top 10 bacteria from STAMP tool. T. harzianum treatment is denoted by blue bar and control by orange bar. The differential abundance between the samples were calculated with G-test (w/Yates’)+Fisher's test for two sample analysis in STAMP tool.
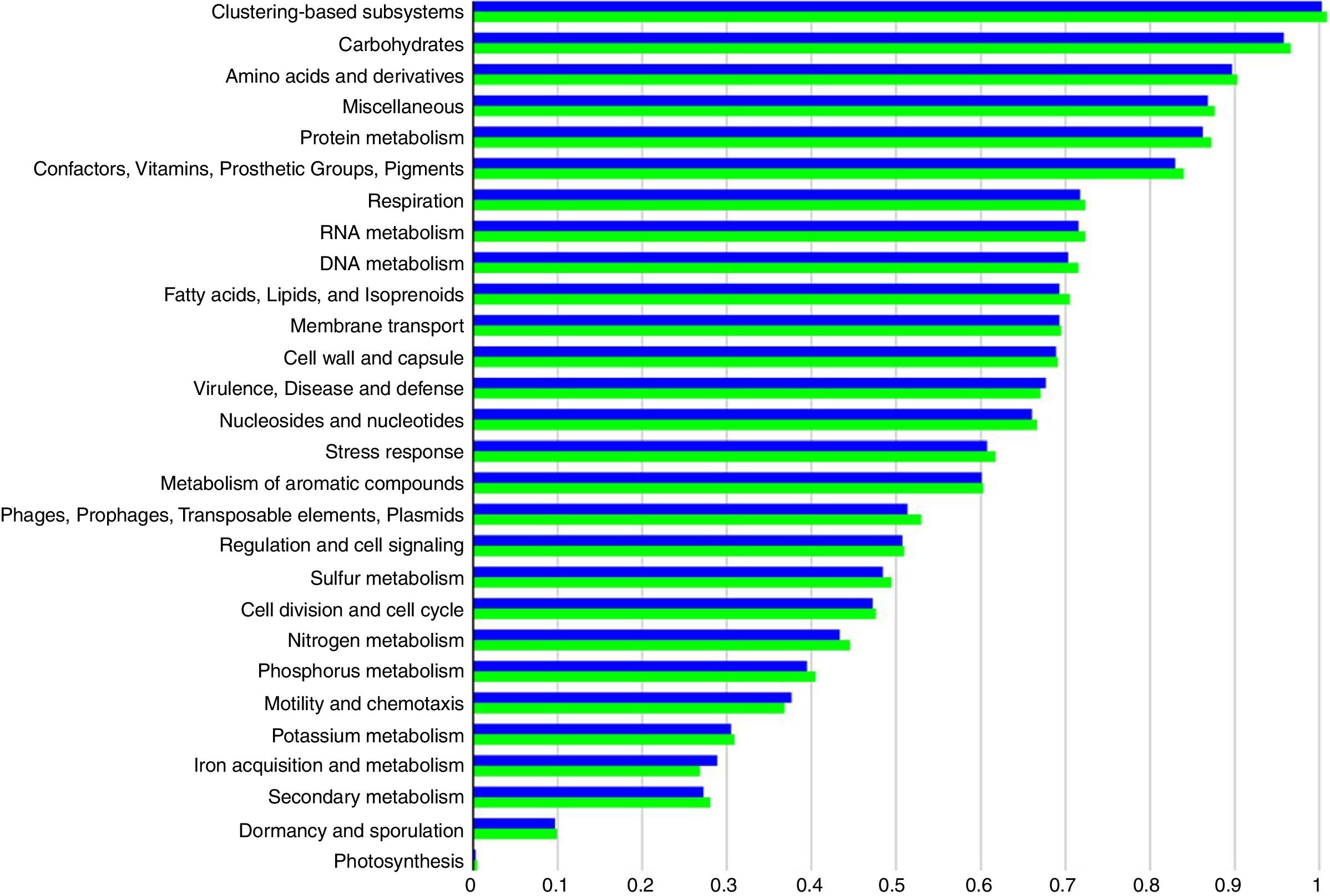
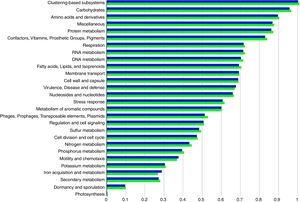
Functional abundance (Fig. 2) between the treatment and control samples using hierarchical classification with subsystem annotation sources showed that rhizosphere in the treatment was with abundant reads for virulence, disease and defense (54,857), motility and chemotaxis (11,992), and ion acquisition and metabolism (8151); while the control recorded 51,271 reads for virulence, disease and defense, 11,564 for motility and chemotaxis, and 7276 for ion acquisition and metabolism.
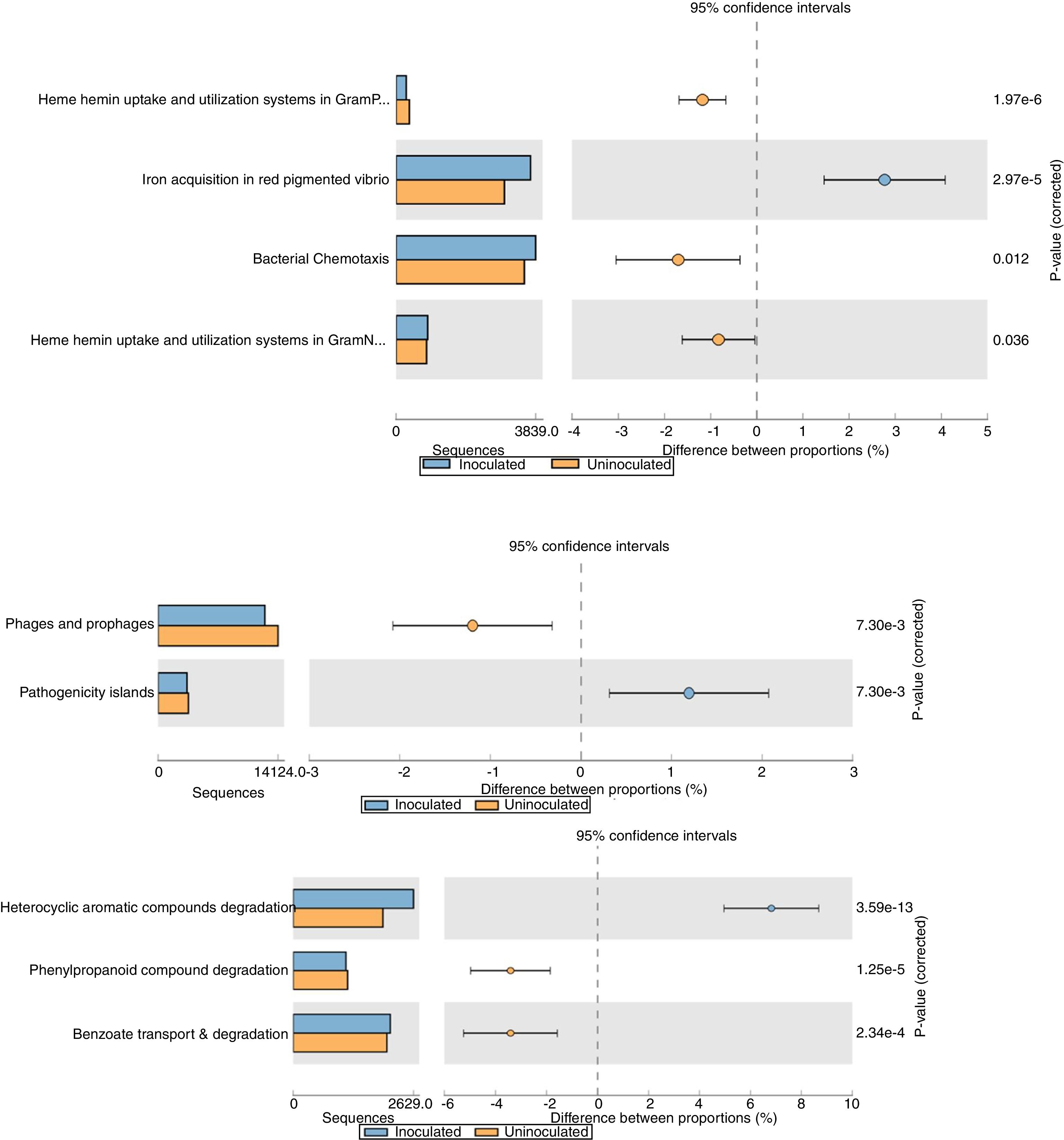
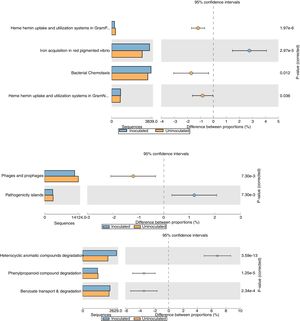
The relative abundance (percentage proportion) for the specific features (iron acquisition and bacterial chemotaxis) from stamp tool analysis is given in Fig. 3. The heme and hemin uptake and utilization systems in Gram negative bacteria (p=0.036) and iron acquisition in red pigmented Vibrio (p=2.97e−5) were abundant in treatment metagenome than in control. This indicates that the probiotic application of T. harzianum increased the microbial action for the metabolism and absorption of iron by the plant. The bacterial chemotaxis was higher in treated sample (p=0.012), which shows the active/increased interaction of rhizosphere microbes on the black pepper roots by the application of T. harzianum. The treated sample recorded reduced abundance on pathogenicity islands, phages and prophages (p=7.30e−3) (Fig. 3).
Functional level extended error bar chart profile for iron acquisition and chemotaxis, phages and prophages, pathogenecity islands and heterocyclic aromatic compounds degradation from STAMP tool. T. harzianum treatment is denoted by blue bar and control by orange bar. The differential abundance between the samples were calculated with G-test (w/Yate's)+Fisher's test for two sample analysis in STAMP tool.
The reduction of pathogenicity island and phages in treatment, when compared to control, provides strong evidence for the selective community recruited by the T. harzianum toward the beneficial use in the cultivation system of black pepper. Though metagenome of control sample showed higher abundance (reads) globally for other functional category (Fig. 2), specific features were observed at the highest functional distribution classification in treatment, which includes metabolism of aromatic compounds, viz., benzoate transport and degradation (p=2.34e−4) and degradation of heterocyclic aromatic compounds (p=3.59e−13). The increased abundance for these metabolism of aromatic compounds brings out that the probiotic application of T. harzianum in black pepper is capable of creating the unique community for the phytoremediation.
DiscussionThe prime objectives of this study was to assess the community changes at the rhizosphere of black pepper pursuant to the inoculation of T. harzianum, and also to unveil the significant effects of T. harzianum on the selective recruitment of specific microbes, and their functional assignments in rhizosphere of black pepper. The results clearly showed that T. harzianum significantly influenced in the selective abundance of beneficial bacteria and fungi, and subsequent growth promotion in black pepper; and the impact at functional level was identified as increased bacterial chemotaxis, virulence, disease and defense, ion metabolism. From the results, increase in the growth parameters, viz., fresh root, fresh shoot, dry root, dry shoot, leaf area index, height of the plant reveals the growth promotion activity of T. harzianum in black pepper, as indicated by other authors too. Anandaraj and Sarma14 reported that the application of T. harzianum (MTCC 5179) resulted in enhanced growth in black pepper with increased number of nodes, and consequently the number of cuttings. Sibi13 also showed the positive influence of T. harzianum (MTCC 5179) on the improvements in the formation of fresh root and shoot, followed by increase in the dry weight of root and shoot in black pepper. Treatment with T. harzianum (MTCC 5179) individually imparted better growth promotion and disease suppression than that of a consortia of plant growth-promoting rhizobacteria alone or in combination with T. harzianum (MTCC 5179).13 These studies indicated growth promotion and the organism was recommended as a component of integrated disease management and without a clear understanding of other mechanisms. The present study unravels the underlying microbial dynamics and major functional processes.
Though the population abundance of bacteria, archea and eukaryote were a little less in treatment than in control, it showed selective abundance (more percentage proportion) of bacteria, viz., A. bacterium and C. koribacter versatili – out of top 10 bacterial species; these bacteria belong to the phylum Acidobacteriaceae, the avid colonizer of the rhizosphere with potent rhizosphere competence.26A. bacterium is capable of growing on diverse collection of complex organic compounds including xylan, cellulose, methyl cellulose, syringate, pectin and ferulate.27Candidatus sp. contains abundance of carboxylase active enzymes (CAZyme) family and are involved in the breakdown, utilization and biosynthesis of diverse structural and storage polysaccharides and resistance to fluctuating temperature and nutrient deficient conditions.28 This selective abundant recruitment of these beneficial bacteria in the treatment might be the major impact for the growth promotion activity by the active breakdown of complex organic compounds by these organisms, thereby creating microclimates for the colonization of microbes in the roots and subsequent interaction with the communities at the rhizosphere. Further, the analysis of black pepper root exudates and action of these bacteria on the root metabolites would give the specific role of these bacteria at the rhizosphere of black pepper.
Unlike in control, the metagenome of treatment showed abundant reads of the beneficial fungi, viz. F. oxysporum, Talaromyces sp. Pestalotiopsis sp. and T. harzianum; a positive correlation with expected beneficial activities as pointed out by different authors: Eparvier and Alabouvette29 showed that increased population of F. oxysporum was better for the biocontrol and disease suppression activity in Flax; many isolates of Talaromyces spp. were shown to promote plant growth.30 Elegant studies have demonstrated that T. flavus antagonizes plant pathogenic fungi.31,32 In present study, higher abundance of the species of Fusarium and Talaromyces in treatment indicates the ecological significance on their population abundance driven by the addition of T. harzianum toward the fitness of black pepper growth and subsequent yield.
Rajan et al.15 showed the biocontrol and disease suppression activities of T. harzianum (MTTC 5179) in black pepper against foot rot disease at field conditions; which was found to be efficiently proliferating in the soil and remained in the soil for long time, apart from imparting protection to the root system against P. capsici. In the present study, the metagenome analysis was performed after four months of treatment, and proved that T. harzianum (MTCC 5179) was able to remain in soil for a long time. Interestingly, the proportion of R. irregularis was higher in the control than in treatment, which indicates the interaction of Trichoderma with the native Vesicular Arbuscular Mycorrhiza (VAM) and modulation of its population. The spore germination and hyphal growth of G. mosseae was stimulated by T. harzianum with the production of volatile compounds.33 In present study, the less abundance of Arbuscular mycorrhizal fungi (AMF) in treated soil might be due to the stimulated growth of AMF by the community recruited by T. harzianum thereby increased colonization inside the plant34 rather than their physical presence in the rhizosphere and vice versa in control. Application of T. harzianum improved better growth of black pepper, which was at par with T. harzianum in combination with AMF. The treatments with AMF alone and in combination with Pseudomonas sp. failed to enhance the growth.13P. fici, an endophyte of tea produces bioactive metabolites and natural products,35 and the analyses of its genome and transcriptome showed that it harbors efficient genes responsible for the synthesis of various secondary metabolites.36 Further functional analysis of the reads on P. fici, from the present metagenome data would give significant insight into its role on black pepper through interaction at rhizosphere.
The metagenome of the treatment in the present study showed higher abundance for iron acquisition and metabolism in red pigmented Vibrio, coupled with heme and hemin uptake and utilization systems in Gram negative bacteria than control; which evidences the influence of T. harzianum in rhizosphere–microbe interaction. Rhizosphere microbiome facilitates the uptake of specific trace elements such as iron. Iron in soil, exists primarily in the insoluble ferric oxide form, which is not available for microbial growth. Based on the scarcity of available irons as well as the toxicity of free iron at elevated concentrations in the environment, bacteria employ a variety of mechanisms to regulate the intracellular iron concentrations.37 On the other hand, plants also play crucial role in increasing the solubility of inorganic iron in the rhizosphere, which may be due to the interaction with microbiome.38 Rhizobacteria are generally motile, and the motility is either random or chemotactic for interacting with the plants.39 In fact, the bacterial chemotaxis was found as abundant in treatment than in control, suggesting that the probiotic application of T. harzianum in black pepper would enable active interaction of the recruited bacterial community in the root system. Anatomical data from the treatment and control also provide ample evidences for the aforesaid inference.34 The abundance of reads on pathogenicity islands, functionality of phages and prophages were found to be less in treatment than in control. The less abundance of human pathogenic fungi as evidenced from the analysis of taxonomy abundance is highly related to the results of functional analysis, which suggests the beneficial effect of probiotic application of T. harzianum, especially in the context of human health.
Rhizoremediation is a specific form of phytoremediation involving plants and their associated rhizospheric microorganisms (bacteria and fungi). Rhizoremediation can either occur naturally or could be facilitated by inoculating soil with microorganisms capable of degrading environmental contaminants. The plant associated non-pathogenic endophytic and the rhizospheric bacteria are the major players in the degradation of toxic metabolites present in soil.40 Heterocyclic aromatic compounds and benzoates are toxic compounds persist for a long time in soil, that leads to ill effects in animals and humans. In the present study, metagenome of treatment recorded higher abundance of reads for the degradation of heterocyclic aromatic compounds, benzoate transport and its degradation. This information would give the positive impact of T. harzianum in the cropping system of black pepper. Further, the functional metagenomics would give more information on bacteria involved in the rhizo remediation through the rhizoecosystem in black pepper.
In conclusion, the population dynamics and functional richness of rhizosphere ecosystem in black pepper influenced by the treatment with T. harzianum provides the ecological importance of T. harzianum in the cultivation of black pepper. On the basis of the present report and previous studies on effect of T. harzianum in the fitness of black pepper; it can be suggested that as mycorhizosphere, another microecological niche, viz., ‘trichorhizosphere’ is also coexists in altering the community dynamics of bacteria and soil fungi; and thus, the rhizosphere microecosystem developed by T. harzianum might contribute a pivotal role in imparting plant health, which is unlike the lone effect of T. harzianum. The methods employed in this study show a significant step toward possible implementation of metagenomics for the functional elucidation of T. harzianum – the valuable biocontrol, growth promoting fungus in the production system of black pepper. The rhizosphere and the trichorhizosphere metagenomes of black pepper elucidated in this study would become important factors in developing any IDM modules in the root ecosystem of black pepper. Further, targeted studies based on the present metagenomic read on each organism and at each component would give enormous information on this microclimate.
Conflicts of interestThe authors declare that there exists no conflict of interest.
This study was funded by Indian Council of Agricultural Research, India, through outreach project “PhytoFuRa (Phytophthora, Fusarium and Ralstonia diseases of Horticultural and Field Crops”. www.phytofura.net.in). The sequencing service was hired from Scigenome, Kochi, Kerala. The authors acknowledge Drs. Devasahayam and Sasikumar, ICAR – Indian Institute of Spices Research, Kozhikodeode, Kerala for their support in carrying out the experiments. PU is grateful to Drs. V. Srinivasan, Hamza, SJ Eapen, K. Kandiannan and D. Prasath, ICAR – Indian Institute of Spices Research, Kozhikode, Kerala for their valuable suggestion in implementing this work.