Fifty seven soil-borne actinomycete strains were assessed for the antibiotic production. Two of the most active isolates, designed as Streptomyces ST-13 and DK-15 exhibited a broad range of antimicrobial activity and therefore they were selected for HPLC fractionation against the most suppressed bacteria Staphylococcus aureus (ST-13) and Chromobacterium violaceum (DK-15). LC/MS analysis of extracts showed the presence of polyketides factumycin (DK15) and tetrangomycin (ST13). The taxonomic position of the antibiotic-producing actinomycetes was determined using a polyphasic approach. Phenotypic characterization and 16S rRNA gene sequence analysis of the isolates matched those described for members of the genus Streptomyces. DK-15 strain exhibited the highest 16S rRNA gene sequence similarity to Streptomyces globosus DSM-40815 (T) and Streptomyces toxytricini DSM-40178 (T) and ST-13 strain to Streptomyces ederensis DSM-40741 (T) and Streptomyces phaeochromogenes DSM-40073 (T). For the proper identification, MALDI-TOF/MS profile of whole-cell proteins led to the identification of S. globosus DK-15 (accession number: KX527570) and S. ederensis ST13 (accession number: KX527568). To our knowledge, there is no report about the production of these antibiotics by S.globosus and S. ederensis, thus isolates DK15 and ST13 identified as S. globosus DK-15 and S.ederensis ST-13 can be considered as new sources of these unique antibacterial metabolites.
Since people started suffering from diseases caused by infectious microorganisms, the quest for their remedies led to the discovery of a large number of antibiotics from microorganisms.1 Most of the research was dedicated to study the polyketide bioactive compounds.2 One of the most important and well acknowledged group of the prolific producers of polyketide antibiotics are the actinomycetes, mainly terrestrial representatives of the genus Streptomyces.3 Polyketides have played an important role in the antibiotic drug discovery with most antibacterial drugs being derived from a natural product or natural product lead.4 Examples of natural products with the polyketide structure include the tetrangomycin and factumycin. Tetrangomycins belong to the group of polyketide angucyclinones, naturally occurring benzaanthraquinones that had been isolated from Streptomyces rimosus and Streptomyces griseus.5–8 Angucyclinones, benz[a]anthraquinone derivatives isolated from Streptomyces spp. have attracted much attention due to their broad and strong biological activities, mainly antiviral, antifungal, and antitumor.5,9–10 Immunosuppressive function of tetrangomycin by reducing the transcription of cytokine genes and the inhibitory activity of tetrangomycin against methicilin resistant Staphylococus aureus (MRSA) was described by Özakin et al.11 Factumycin is a linear polyketide elfamycin compound and had been isolated from Streptomyces lavendulae and Streptomyces griseus.12–14 According to Thaker et al.15 a screen of factumycin in combination with known antibiotics revealed unexpected synergy with tetracyclines, offering a possible new application of factumycin as a lead in Gram-negative targeted antibiotic combination therapy. Identification of factumycin antibiotic has high importance, because Gram - negative pathogens are becoming increasingly antibiotic resistant and consequently a serious clinical challenge.15
Therefore, an attempt has been made to isolate the antibiotic-producing actinomycetes with emphasis on the selection and the detailed identification of Streptomyces polyketide-producers.
Material and methodsSoil samples and isolation of actinomycetesActinomycetes were isolated by the dilution agar plating method from the forest soils located in Vietnam, Phu Quoc National Park (10°09¿33.49“S 103°59¿07.02”V) and in Sardinia, Foresta di Monte Pisanu (40°25′30.24“S 8°58′34.72″V). An aliquot of 0.1ml of solution was taken and spread over the surface of starch-casein medium.16 Plates were incubated at 30°C and monitored after 7 days. The selected strains were maintained by cultivation on the ISP2 medium17, incubated at 30°C for 10 days and stored at – 20°C in the presence of glycerol (30% v/v). For all acquired actinomycete isolates, 16S rRNA sequences as well as microscopic (shape of spore chains) and macroscopic morphology (colors of aerial and substrate mycelium, soluble pigments and melanin) were studied in accordance with the guidelines established by the International Streptomycete Project.17
Screening for antimicrobial activityThe morphologically different actinomycete isolates were screened against four Gram - positive bacteria [Bacillus subtilis (DSM 10), Micrococcus luteus (DSM1790), Staphylococcus aureus (Newman), Mycobacterium smegmatis (ATCC 700084)], four Gram - negative bacteria [Escherichia coli (DSM 1116), Escherichia coli (TolC), Pseudomonas aeruginosa (PA14), Chromobacterium violaceum (DSM 30191)], two yeasts [Candida albicans (DSM 1665), Pichia anomala (DSM 6766)] and one filamentous microscopic fungus Mucor hiemalis (DSM 2656) obtained from DSMZ (German Collection of Microorganisms and Cell Cultures), Braunschweig, Germany and ATCC (American Type Culture Collection), Manassas, VA 20110, USA. Raw extracts were prepared from 20ml of 5 day old liquid culture using ethyl acetate (Sigma-Aldrich, USA) according to Wink (2014). Ethylacetate was evaporated and extract was dissolved in 1ml of ethylacetate: acetone: methanol (1:1:1) solution. Detection of biological activity was carried out by preparing 4 - 6h cultures of indicator bacteria followed by dilution with Mueller- Hinton (MH) broth (Merck, Germany) and yeast and fungi by dilution with Mycosel broth18 to obtain 0.01 McFarland turbidity. The antimicrobial activity of streptomycetes was performed by using the broth micro - dilution method 19 in 96 - well microplates (BRAND, Germany). The dose in well A corresponds to the extract made from 1.33ml Streptomyces liquid culture per 1ml of the inoculated broth. Concentration decreased 2 fold in wells B-H. The readings were made after 24h of incubation through visual observation of the growth.
HPLC and LC/MS analysis of selected crude extractsThe most active raw extracts were selected for the fractionation by liquid - chromatography (Agilent 1100) equipment. The wavelength monitoring was performed at 210 and 360nm. Sample (5μl) was injected onto a HPLC column (X-Bridge 3.5μm, 2.1x100mm; Waters, Milford, USA). Separation was performed using the buffers A2: 950ml H2O, 50ml acetonitrile + 0.05mM (385mg.l-1) ammonium acetate + 40μl acetic acid; B2: 50ml H2O, 950ml acetonitrile, 0.05mM (385mg.l-1) ammonium acetate + 40μl acetic acid and a DAD detector (200-400nm). Fractions (0.15ml) from the HPLC column were collected in a 96 well plates every 0.5min and dried with heated nitrogen in MiniVap (Porvair Sciences, UK) for 45-60min at 40°C. Afterwards, each well was filled with 150μl of the selected formerly inhibited test microorganisms and incubated at 30°C for 24h. Due to the fact that inhibited wells were visible, the extracts were applied to an LC–MS system [(Agilent 1200 series with DAD detector (200–600nm) in connection with a maXis UHR–TOF mass spectrometer (Bruker Daltonics, USA)] for peak–activity correlation. Samples were analyzed using a Waters ACQUITY UPLC BEH C18 Column, 2.1 x 50mm, 1.7μm. The mobile phase consisted of H2O with 0.1% formic acid as solvent A and CH3CN with 0.1% formic acid as solvent B at a flow rate of 0.6ml.min-1. The gradient was as follows: 0.5min 5% B, 19.5min 95% B and 10min 95% B. Equilibration time between samples was 5min. The column temperature was maintained at 40°C. The main software for processing of results was Data Analysis included in the Compass-software from Bruker (USA).
Genomic DNA isolation, PCR conditions and phylogenetic analysis of the most active isolatesFor the taxonomic identification, the 16S rRNA gene of the selected streptomycetes was sequenced. The isolation of genomic DNA was done by the method described by Sambrook et al. 20 and amplified by PCR using primers F27 and R1492 according to Cook and Meyers.21 The reaction mixture was made in a total volume of 50μl (5μl of 10 × DreamTaq Green PCR buffer, 5μl of 2 mmol.dm -3 dNTP, 2μl of each 10μmol.dm-3 primer, 0,3μl Taq DNA polymerase and 0,5μl of template DNA (approximately 20 ng)). The PCR reaction ran in the thermo cycler Biometra T Personal (Germany) under the following conditions: 95°C for 3min, 40 cycles of 95°C for 30sec, 56°C for 30sec, 72°C for 90sec and final extension at 72°C for 10min. The PCR products were purified with Exonuclease I and Thermosensitive Alkaline Phosphatase (Sigma-Aldrich, USA).
For a rough classification, the 16S rRNA genes were sequenced in MacroGen Company, South Korea. The obtained 16S rRNA sequences were checked for quality and assembled using the program SeqMan II. The similarity and homology of the 16S rRNA gene sequences were analyzed with the similar existing sequences available in the data bank of NCBI using BLAST search (http://www.ncbi.nlm.nih.gov/BLAST). The phylogenetic tree was constructed with the Maximum - Likelihood method22 using PhyML23, with bootstrap values based on 1000 replications.
Cultural and biochemical characteristicsFor the taxonomic identification, additional to the 16S rRNA gene sequences and morphological traits, physiological and biochemical features of the isolates DK15 and ST13 were determined. Physiological criteria such as a sodium chloride resistance was tested on ISP9 medium supplemented with 2.5, 5, 7.5 and 10% of NaCl, utilization of different carbon sources (glucose, mannitol, arabinose, inositol, mannose, fructose, galactose, rhamnose, sucrose, xylose) was tested on ISP9 medium with the final concentration of carbon sources adjusted to 1%, pH (levels of 2-9) and temperature (4, 25, 28, 30, 37 and 42°C) tolerance was tested on the ISP2 medium. Commercially available test kits such as an ApiZym® and ApiCoryne® (bioMérieux, France) were used for the biochemical characterisation. The Api stripes were inoculated following by manufacturer's manual. The observed characteristics were compared with the phylogenetically related type cultures obtained from German Collection of Microorganisms and Cell Cultures (DSMZ), Braunschweig, Germany.
MALDI-TOF MS identificationFor the MALDI–TOF MS analysis, cellular proteins were extracted according to Loucif et al. 24. For both strains, four extractions were performed, and 1μl of each suspension was deposited on the MALDI–TOF steel target plate (Bruker Daltonics, Germany) in three replicates. The plate was allowed to dry at room temperature and then overlaid with 1μl of matrix solution containing α–cyano–hydroxycinnamic acid (SigmaAldrich, USA) saturated with acetonitrile (50%), trifluoroacetic acid (2.5%), and ultrapure water. This mixture was allowed to co–crystallize with the samples. Analysis of protein spectra was performed with the Microflex LT MALDI–TOF MS spectrometer. The measurements were taken in the linear positive ion mode with voltage of 20kV over a mass range of 2–20 kDA. The resulting spectra were analyzed using MALDI Biotyper, and flexAnalysis (Bruker Daltonics). The obtained spectra were compared with our own created database from type Streptomyces cultures obtained from DSMZ, Braunschweig, Germany, which included all the most phylogenetically related species. For the clustering of the Streptomyces species, a mean spectra projection (MSP) dendrogram was constructed with MALDI Biotyper 3.1.
ResultsAntimicrobial activity of the crude extracts and identification of the antibacterial compounds by LC/MS analysis
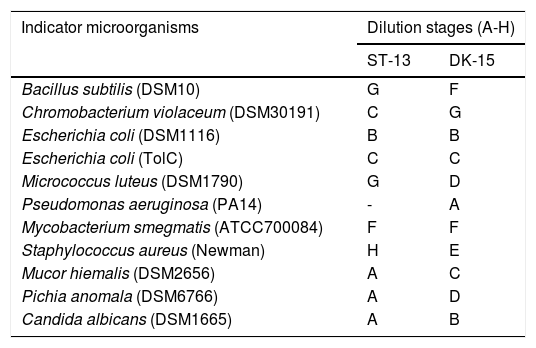
To establish effective bacteria for the control of human pathogens, a total of 57 actinomycetes were isolated from the forest soil samples. 28 isolates representing 10 species were acquired from Vietnam soil while 29 isolates of 8 species were found in Sardinia soil. Out of all 57 isolates, 8 did not show any antimicrobial activity, 13 low activity (A-B dilution of crude extract), 34 high (C-F), and 2 very high (G-H) based on the results from a classical approach of assessing microplates for the growth inhibition. Two of the most active strains were designed as ST13 and DK15. As shown in Table 1, the crude extracts exhibited high antimicrobial activities against Gram - positive bacteria and against Gram-negative bacterium Chromobacterium violaceum DSM30191, but weak activity against Escherichia coli DSM1116 and Pseudomonas aeruginosa PA14.
Dilution stages of the crude extracts prepared from isolates DK-15 and ST-13.
| Indicator microorganisms | Dilution stages (A-H) | |
|---|---|---|
| ST-13 | DK-15 | |
| Bacillus subtilis (DSM10) | G | F |
| Chromobacterium violaceum (DSM30191) | C | G |
| Escherichia coli (DSM1116) | B | B |
| Escherichia coli (TolC) | C | C |
| Micrococcus luteus (DSM1790) | G | D |
| Pseudomonas aeruginosa (PA14) | - | A |
| Mycobacterium smegmatis (ATCC700084) | F | F |
| Staphylococcus aureus (Newman) | H | E |
| Mucor hiemalis (DSM2656) | A | C |
| Pichia anomala (DSM6766) | A | D |
| Candida albicans (DSM1665) | A | B |
Dilution stages represented stepwise 2-fold dilution of crude extracts (A – crude extract from 1,33ml of Streptomyces culture in 1ml of inoculated media, B – 0.66ml.ml-1; C – 0.66ml.ml-1; D – 0.33ml.ml-1;E – 0.17ml.ml-1; F – 0.08ml.ml-1; G – 0.04ml.ml-1;and H – 0.02ml.ml-1)
Although the nature and type of the active antibacterial compounds were not clear, the prominent antibacterial activities of isolates highlight them as candidates for further investigation. Therefore both ethylacetate extracts were subjected to HPLC fractionation followed by LC/MS analysis of the active compounds. The range of inhibited wells for selected strains revealed that the tested extracts exhibited the strongest antimicrobial activity against Gram-positive bacterium Staphylococcus aureus (inhibited wells until H, strain ST-13) and Gram-negative bacterium Chromobacterium violaceum (inhibited wells until G, strain DK-15).
The peak-activity-correlation test (Figure 1) of ST-13 extract revealed that the active fractions after HPLC fractionation were located at retention times 13.0-13.5min. The dominant peak in HPLC chromatogram appearing at this retention time exhibited UV-VIS maxima at 268nm and ESI-HRMS spectrum showing the prominent ion clusters for [M+H]+ at m/z 339.0861. These data correlated to tetrangomycin.
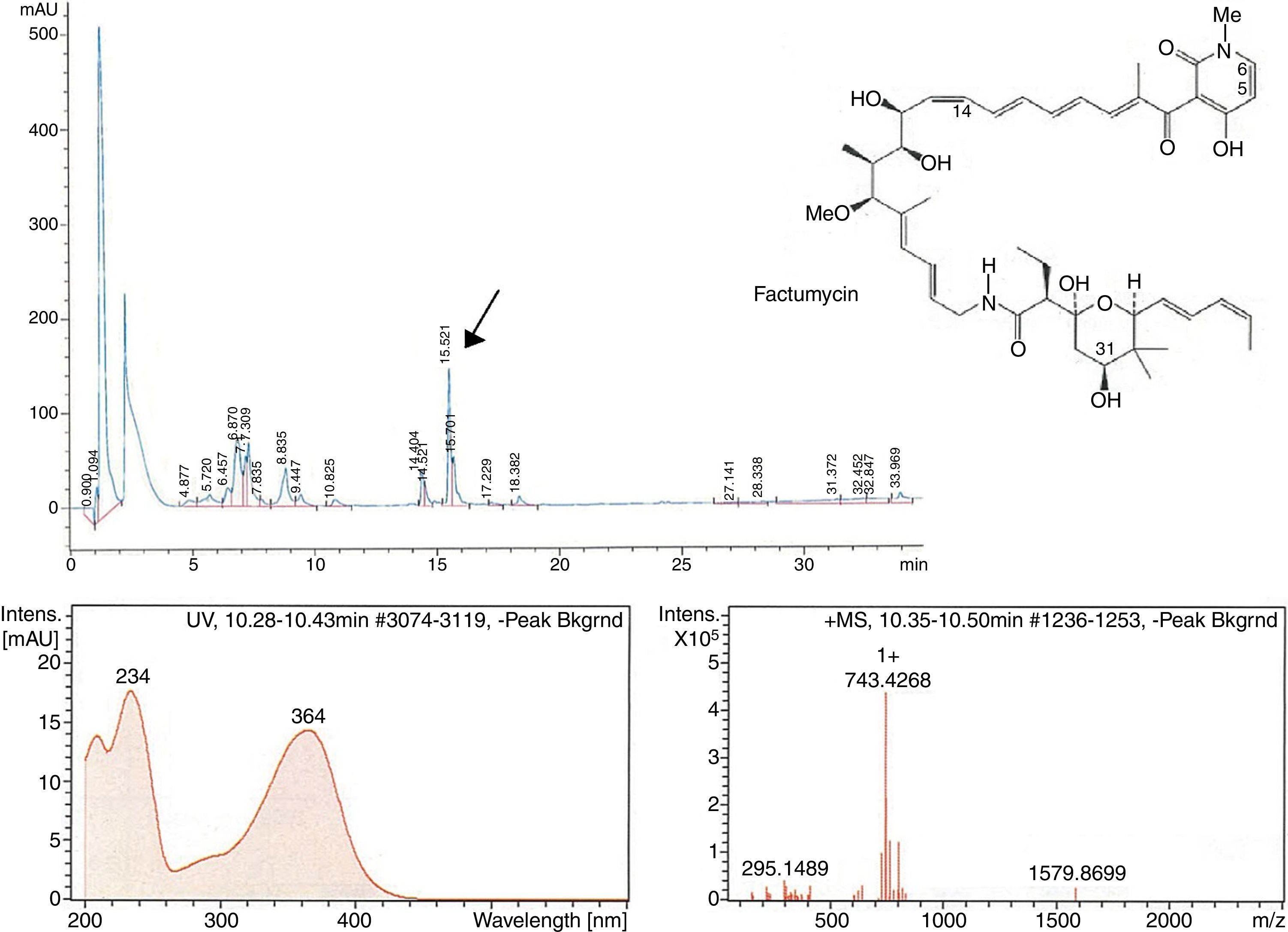
The second peak-activity-correlation test (Figure 2) of DK-15 extract revealed that the active fractions were located between retention times 16.0-16.5min. The peak appearing at this retention times exhibit UV-VIS maxima at 364nm and ESI-HRMS spectrum showing the prominent ion clusters for [M +H]+at m/z 743.4268. These data correlated to factumycin.
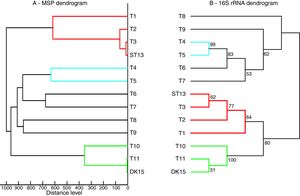
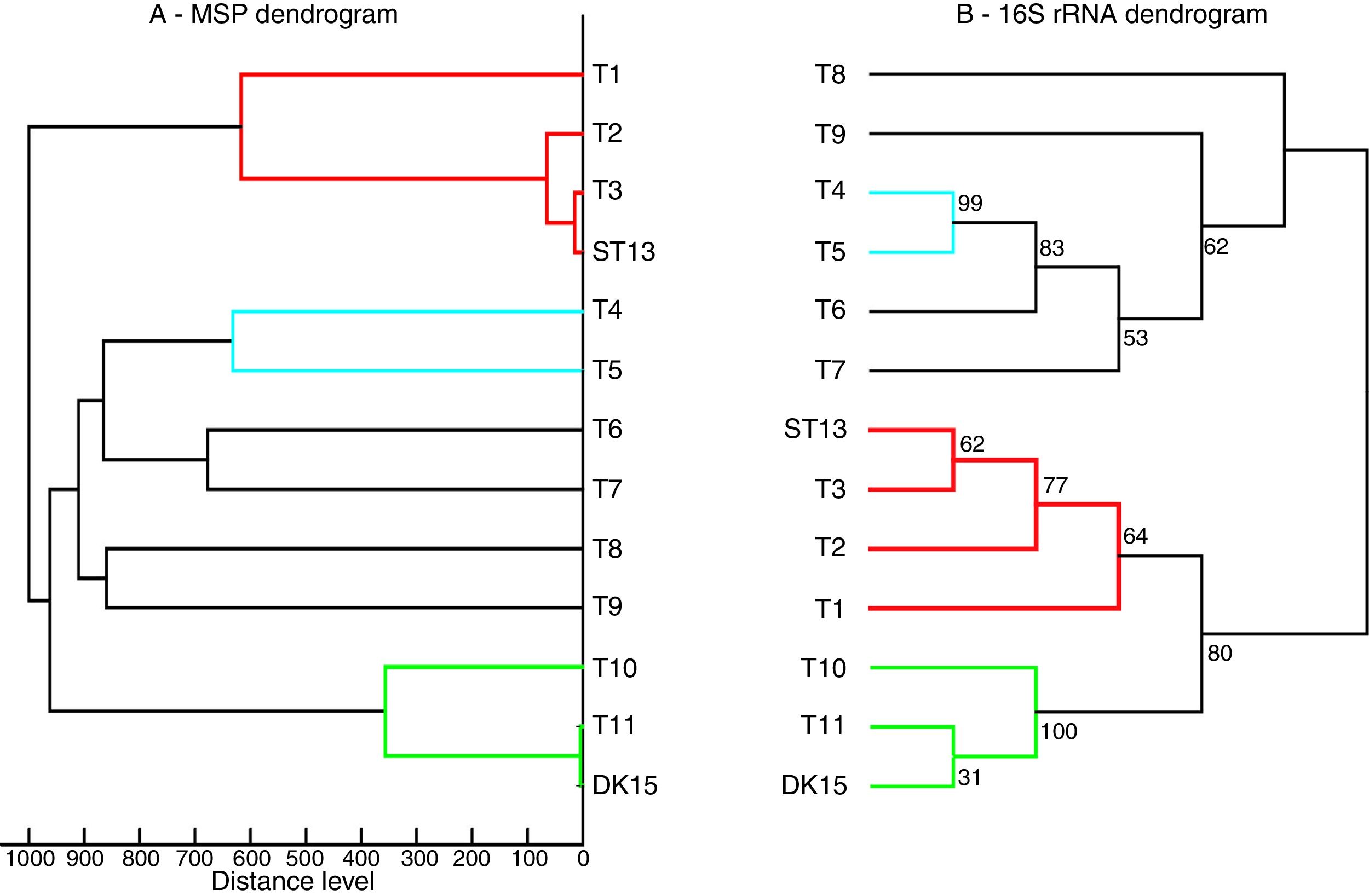
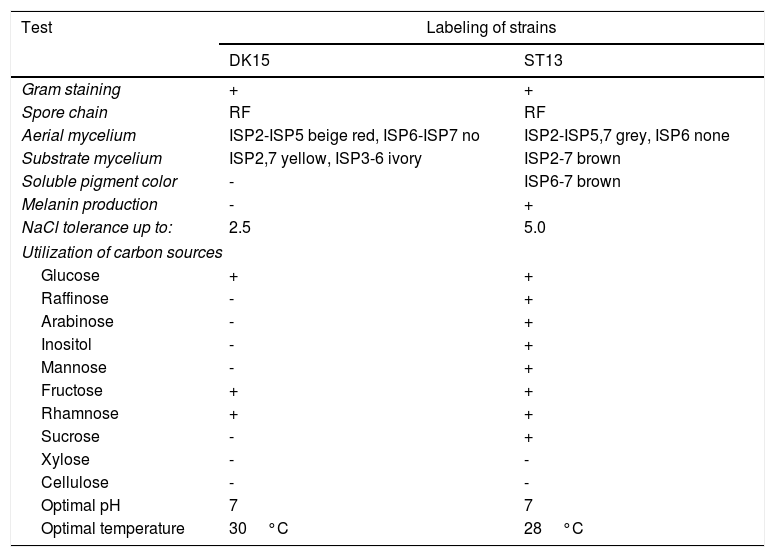
Both strains were observed to be Gram-positive, aerobic, non-motile and with abundant substrate and aerial mycelium typical for the genus Streptomyces. In the present study, isolates DK15 and ST13 were subjected to the molecular, protein and polyphasic identification. The 16S rRNA genes were sequenced and compared with the 16S rRNA sequences of previously described the most related type cultures. A comparison of the sequences using Genbank database showed the highest similarity of ST13 with Streptomyces ederensis DSM-40741T (99%, 1450 of 1460 bases were identical, 2 gaps) and Streptomyces phaeochromogenes DSM-40073T (99%, 1448/1463, 4 gaps). Isolate DK15 showed identical sequences with Streptomyces globosus DSM-40815T and Streptomyces toxytricini DSM-40178T. A maximum likehood phylogenetic analysis confirmed these findings (Figure 3). The sequences of 16S rRNA gene were deposited under accession numbers KX527570 for DK15 and KX527568 for ST13. The MSP dendrogram showed high concordance with the clustering and topology of the 16S rRNA gene tree, and clustered strain ST-13 with Streptomyces ederensis (DSM-40741T) and strain DK-15 with Streptomyces globosus (DSM-40815T) (Figure 3). These results were in concordance with cultural and physiological features of the tested strains (Table 2) and the most related type strains (data available in Compendium of Actinobacteria).25
Comparison between 16S rRNA phylogenetic tree and the MSP dendrogram. A. Mean spectra projection (MSP) dendrogram, B. 16S rRNA based phylogenetic tree of Streptomyces type strains, T1-T11 – DSM type cultures used for grouping of Streptomyces species, T1 - Streptomyces viridis DSM-42078 (T), T2 - S. phaeochromogenes DSM- 40788 (T), T3 - S. ederensis DSM-40741 (T), T4 - S. iakyrus DSM-40482 (T), T5 - S. griseochromogenes DSM-40499 (T), T6 - S. gougerotii DSM-40324 (T), T7 - S. albus DSM-40313 (T), T8 - S. diastaticus DSM-40495 (T), T9 - S. alboflavus DSM-40045 (T), T10 - S. toxytricini DSM-40178 (T), T11 - S. globosus DSM-41122 (T).
Cultural, morphological and physiological characteristics of strains DK-15 and ST-13.
| Test | Labeling of strains | |
|---|---|---|
| DK15 | ST13 | |
| Gram staining | + | + |
| Spore chain | RF | RF |
| Aerial mycelium | ISP2-ISP5 beige red, ISP6-ISP7 no | ISP2-ISP5,7 grey, ISP6 none |
| Substrate mycelium | ISP2,7 yellow, ISP3-6 ivory | ISP2-7 brown |
| Soluble pigment color | - | ISP6-7 brown |
| Melanin production | - | + |
| NaCl tolerance up to: | 2.5 | 5.0 |
| Utilization of carbon sources | ||
| Glucose | + | + |
| Raffinose | - | + |
| Arabinose | - | + |
| Inositol | - | + |
| Mannose | - | + |
| Fructose | + | + |
| Rhamnose | + | + |
| Sucrose | - | + |
| Xylose | - | - |
| Cellulose | - | - |
| Optimal pH | 7 | 7 |
| Optimal temperature | 30°C | 28°C |
+ positive growth, - negative growth, ISP - International Streptomycete Project
Activity of the extracellular enzymes was quantified using the Api Zym® and Api Coryne® stripes. A huge enzymatic potential was discovered within the tested streptomycetes. It was found that isolates showed high alkaline phosphatase, esterase, leucinearylamidase, valinearylamidase, glucosidase, N-acetyl-glucoseamidase and urease activity (> 40 nmol).
From the available literature we found out, that isolate DK15 identified as Streptomyces globosus DK15 represent a new source of factumycin antibiotic and isolate ST13 idenfied as Streptomyces ederensis ST13 represent a new source of tetrangomycin antibiotic. The results from present study indicated, that during our research streptomycete programme we found out two new producers of polyketide antibacterial compounds.
DiscussionUsing crude ethylacetate extracts prepared from ST13 and DK15 cells, the high antimicrobial activities were detected against Gram-positive and Gram-negative bacteria. Chromatographic analysis of crude extract of our strains ST13 and DK15 led to the identification of the antibacterial compounds factumycin and tetrangomycin with polyketide structure.
Polyketides constitute an important group of secondary metabolites highly relevant as therapeutics for clinical use as many of them are potent antibiotic, antifungal, antitumor, immunosuppressant, antiviral or antiparasitic agents.26,27 Many polyketide secondary metabolites are produced by a group of filamentous gram-positive bacteria of the Actinomycetales order.28 Tetrangomycin, one of the first members of angucyclinones, was previously isolated from the culture broth of Streptomyces rimosus, or S. cyanogenus S-136. 7,29 Özakin et al.11 reported high antimicrobial activity of crude extract which contained tetrangomycin from Streptomyces sp. CAH29 against MRSA. Our results also indicate highest activity against S. aureus. Activity of tetrangomycin is probably based on inhibition of the staphyloxanthin production by S. aureus and there is a potential of use this substance in new MRSA drug.
Factumycin were previously found in S. collinus, Streptomyces sp. WAC5292, and S. lavendulae.30,15,13High antimicrobial activity of the factumycin was reported mainly against Gram negative bacteria.15In our case, inhibition of gram negative as well as gram positive species (M. smegmatis, B. subtilis) was detected however it depends on testing strains.
The search for polyketide antibiotics requires the screening and reliable identification of Streptomyces strains which has always been considered to be very difficult31. Despite specificity of 16S rRNA region, there exist some drawbacks and inconsistencies in this method of identification.32 In our study, sequences of 16S rRNA gene did not allow us identification of strain DK15. Recently, study of Loucif et al.24 demonstrated that protein analysis using MALDI-TOF-MS can be used for the rapid identification of Streptomyces species. Comparison of protein spectra together with analysis of 16S rRNA gene and detailed morphological, physiological and enzymatic analysis provide very exact results about taxonomic position of streptomycete strains we analyzed. Strain ST13 was identified as S. ederensis and strain DK15 as S. globosus. Tetrangomycin have not been previously reported from S. ederensis (producer of moenomycin, bambermycin, and phaeochromycin)33-35 and factumycin from S. globosus (producer of actinomycin, and 2-deoxystreptamine aminocyclitol aminoglycoside antibiotic)36, and therefore we reported a new producers of these high active antibacterial compounds. These strains were isolated from the forest soils in national parks. Such soils can be considered as rich deposits of novel active strains with high antimicrobial activity.
Conflict of interestThe authors have not declared any conflict of interest.
The authors are grateful to the Helmholtz Centre for Infection Research (Microbial Strain Collection Group), Braunschweig, Germany for the scholarship and support of the results. This study was also supported by the Scientific Grant Agency of the Ministry of Education of the Slovak Republic and of the Slovak Academy of Sciences grant no. VEGA 1/0305/17. The authors are grateful to Ms. Martina Drexlerova for the editing and English corrections.



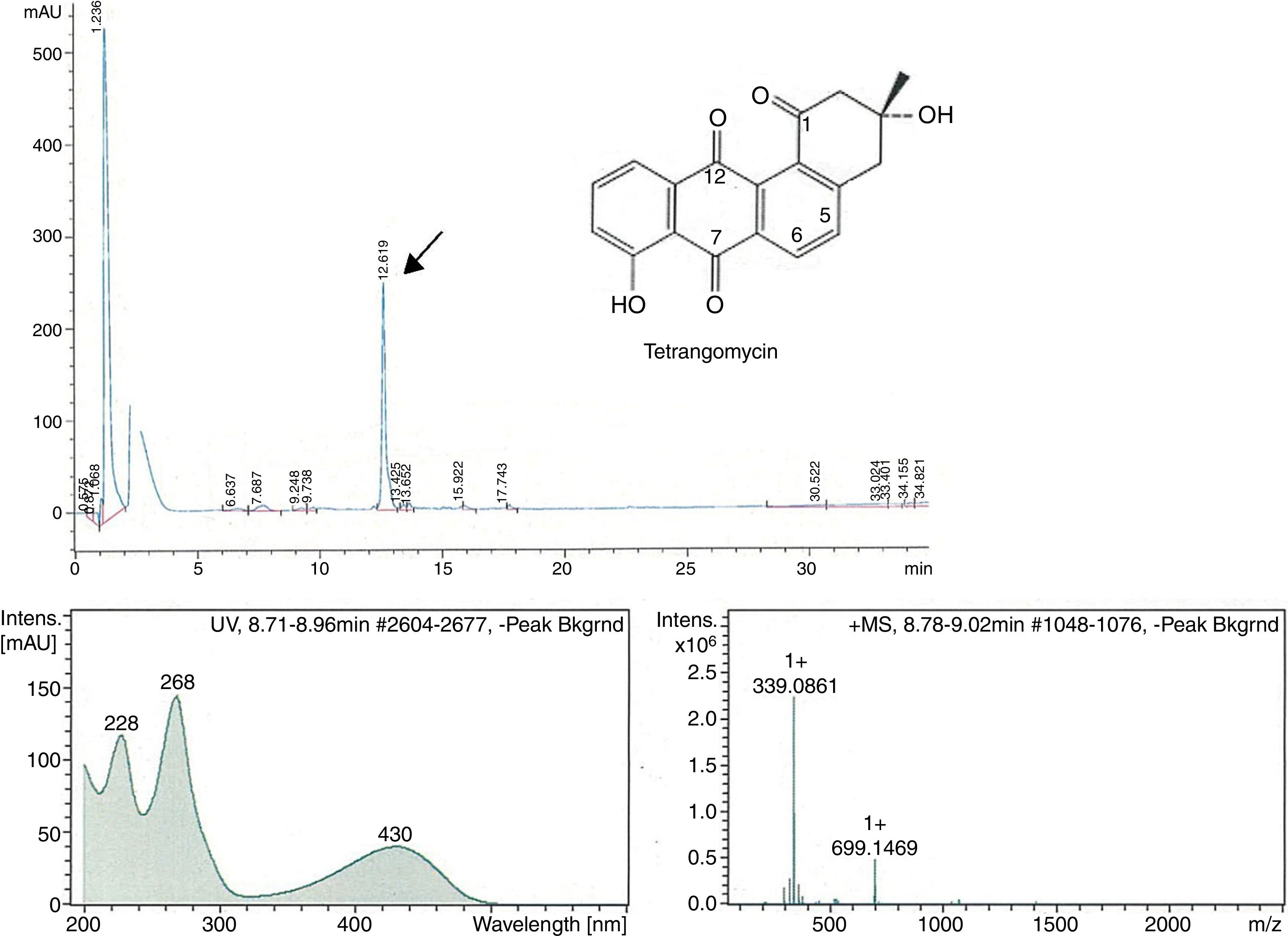
![Fractionation RP-HPLC chromatogram of tetrangomycin, UV spectrum of tetrangomycin with the max at 268nm, and ESI-HRMS spectrum, showing the prominent ion clusters for [M+H]+ at m/z 339.0861. Fractionation RP-HPLC chromatogram of tetrangomycin, UV spectrum of tetrangomycin with the max at 268nm, and ESI-HRMS spectrum, showing the prominent ion clusters for [M+H]+ at m/z 339.0861.](https://static.elsevier.es/multimedia/15178382/0000004900000004/v1_201810050636/S1517838217305518/v1_201810050636/en/main.assets/thumbnail/gr1.jpeg?xkr=ue/ImdikoIMrsJoerZ+w96p5LBcBpyJTqfwgorxm+Ow=)
![Fractionation RP-HPLC chromatogram of factumycin, UV spectrum of factumycin with the max. at 364nm, and ESI-HRMS spectrum, showing the prominent ion clusters for [M+H]+ at m/z 743.4268. Fractionation RP-HPLC chromatogram of factumycin, UV spectrum of factumycin with the max. at 364nm, and ESI-HRMS spectrum, showing the prominent ion clusters for [M+H]+ at m/z 743.4268.](https://static.elsevier.es/multimedia/15178382/0000004900000004/v1_201810050636/S1517838217305518/v1_201810050636/en/main.assets/thumbnail/gr2.jpeg?xkr=ue/ImdikoIMrsJoerZ+w96p5LBcBpyJTqfwgorxm+Ow=)