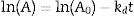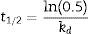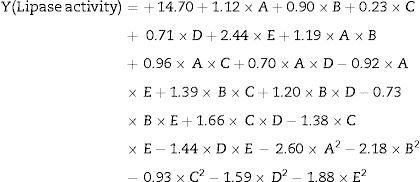The purpose of this study was to isolate, purify and optimize the production conditions of an organic solvent tolerant and thermostable lipase from Acinetobacter sp. AU07 isolated from distillery waste. The lipase production was optimized by response surface methodology, and a maximum production of 14.5U/mL was observed at 30°C and pH 7, using a 0.5% (v/v) inoculum, 2% (v/v) castor oil (inducer), and agitation 150rpm. The optimized conditions from the shake flask experiments were validated in a 3L lab scale bioreactor, and the lipase production increased to 48U/mL. The enzyme was purified by ammonium sulfate precipitation and ion exchange chromatography and the overall yield was 36%. SDS-PAGE indicated a molecular weight of 45kDa for the purified protein, and Matrix assisted laser desorption/ionization time of flight analysis of the purified lipase showed sequence similarity with GDSL family of lipases. The optimum temperature and pH for activity of the enzyme was found to be 50°C and 8.0, respectively. The lipase was completely inhibited by phenylmethylsulfonyl fluoride but minimal inhibition was observed when incubated with ethylenediaminetetraacetic acid and dithiothreitol. The enzyme was stable in the presence of non-polar hydrophobic solvents. Detergents like SDS inhibited enzyme activity; however, there was minimal loss of enzyme activity when incubated with hydrogen peroxide, Tween 80 and Triton X-100. The kinetic constants (Km and Vmax) revealed that the hydrolytic activity of the lipase was specific to moderate chain fatty acid esters. The Vmax, Km and Vmax/Km ratio of the enzyme were 16.98U/mg, 0.51mM, and 33.29, respectively when 4-nitrophenyl palmitate was used as a substrate.
Lipases (EC 3.1.1.3) are enzymes that cleave ester bonds in lipidic substrates. In the presence of water, they catalyze the hydrolysis of triglycerides to form monoglycerides, diglycerides, glycerol and free fatty acids. Lipases are serine hydrolases and are active at the lipid-water interface.1,2 They are ubiquitous in nature and are found in a variety of plants, animals and microorganisms.3 Most bacterial lipases are secreted extracellularly and are versatile biocatalysts that carry out a variety of reactions viz. hydrolysis, esterification, transesterification, inter esterification, acidolysis, and aminolysis.4,5 Lipases utilize a wide spectrum of substrates, and some of them are stable at extreme temperature, and pH conditions and in organic solvents. They are used as catalysts for reactions in reduced water environments.6 Frequently, the substrates of lipases are insoluble in aqueous solution. Hence, conducting the reactions in organic solvents can improve the dissolution of substrates and increase substrate availability, in addition to aiding in the easy separation of enzymes from substrates or products.7,8
Lipolytic strains isolated from industrial effluents show potential utility in biodegradation and bioremediation. The biofilm formed by lipase secreting organisms can be used to degrade fats and oils.9 Therefore, we have produced, purified and biochemically characterized a lipase isolated from Acinetobacter sp. AU07. We also optimized the physical conditions by employing response surface methodology (RSM) to improve lipase production.
Materials and methodsChemicalsEnzyme substrates (4-nitrophenyl esters) and inhibitors were procured from Sigma (St. Louis, USA). Chemicals for media preparation were purchased from Hi-Media (Mumbai, India). The ion-exchange chromatography sorbent diethylaminoethyl (DEAE) Sepharose fast flow was purchased from GE Health care. All chemicals used were of analytical grade.
Isolation and screening of lipase producing organismsThe lipase producing organisms were isolated from a distillery unit. The liquid sample (1mL) was suspended in 9mL sterilized water, serially diluted and spread on selective medium containing sesame oil as the sole carbon source and incubated at 37°C for 24h. This selective medium contained 2.0g/L peptone, 5.0g/L NaCl, 20 (v/v) sesame oil (emulsified with 0.01% Triton X-100), and 15.0g/L bacteriological agar. To screen for lipase production, individual bacterial colonies were streaked onto plates containing tributyrin 1.25g/L (emulsified with 0.01% Triton X-100) and 15g/L bacteriological agar. The plates were incubated at 37°C for 24h, and colonies that formed a zone of clearance were lipolytic positive strains. The thirteen positive isolates were further screened for maximal secretion of extracellular lipase by assaying the lipase activity in liquid culture using 4-nitrophenyl palmitate as a substrate at 37°C. The Acinetobacter sp. AU07 strain, which showed the highest activity, was selected for further study. This strain was maintained in glycerol stocks (50%, v/v) and stored at −20°C.
Identification of the lipolytic strainThe taxonomy of the isolated strain was examined using Bergey's Manual of Determinative Biology and confirmed by 16S rDNA sequencing. A BLAST analysis of the 16S rDNA sequence identified the strain as Acinetobacter sp. AU07. The 800bp 16S rDNA gene sequence of the Acinetobacter sp. AU07 strain has been submitted to the GenBank database, with the accession number HQ914215.
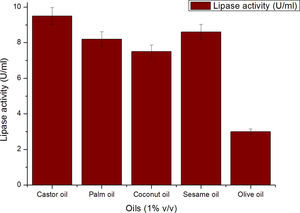
Optimization of media and culture conditions for lipase production by response surface methodologyOne-factor-at-a time strategyThe production of lipase by Acinetobacter sp. AU07 was performed using different vegetable oils as inducers viz. castor oil, palm oil, coconut oil, sesame oil, and olive oil at (1%, v/v), with 1% inoculum at 30°C for 20h in a rotary shaker (150rpm). The media containing oils were emulsified with 0.25% gum acacia and were adjusted to pH 7.0. The individual effects of pH, temperature and inducers were monitored and optimized. The cell-free supernatant was recovered by centrifugation at 12,500×g for 10min at 4°C and used to determine extracellular lipase activity.
The nutrient medium containing castor oil enhanced lipase secretion and was therefore selected for further optimization of lipase production by central composite design (CCD) and response surface analysis. The variables utilized for RSM were as follows: temperature, pH, agitation, inducer concentration, and inoculum volume. All five variables were tested at three levels and designated as −1, 0, +1. The data from CCD were analysed by the least squares method. A total of 26 experiments were conducted. The response values (Y) in each trial were the average of the three replicates. The statistical software package ‘Design Expert’ software (Version 8.0, Stat-Ease Inc., Minneapolis, USA) was used to analyse the experimental designs.
Spectrophotometric assay for lipase using 4-nitrophenyl palmitateThe lipase activity was measured spectrophotometrically using 4-nitrophenyl palmitate (4-NPP) as the substrate. First, 400μl of the enzyme was equilibrated with 50μl of 1M Tris–HCl buffer (final concentration 50mM), and diluted with 530μl sterile water. Next, 20μl of 4-NPP (final concentration 1mM) was added, and the reaction was incubated at 30°C for 10min. The concentration of released 4-nitrophenol was measured at 410nm. The protein content was determined by Bradford's method using the Bio-Rad assay reagent, 10 with bovine serum albumin (BSA) as a standard.
Purification of lipaseThe culture was grown for 16h and centrifuged at 12,500×g for 15min at 4°C. To precipitate the proteins the culture supernatant was added with solid ammonium sulfate (60% saturation) with continuous stirring and incubated at 4°C for 24h. After incubation, the sample was centrifuged at 4°C for 15min at 12,500×g, and the resulting pellet was dissolved in 10mM Tris–HCl buffer (pH 8.0) and dialyzed with the same buffer for 12h at 4°C. The dialyzed protein sample was then loaded on-to a DEAE Sepharose anion exchanger, which had been pre-equilibrated with 20mM Tris–HCl and eluted with a buffered NaCl (0.1–1M) concentration gradient. The lipase containing fractions were pooled and dialyzed with 10mM Tris–HCl buffer. The resulting dialyzed protein sample was then lyophilized.
RP-HPLC analysis of purified lipaseThe purity of the enzyme was analysed by RP-HPLC using an Agilent 1100 HPLC system with a C-18 column (Zorbax C-18, 4.6mm×250mm i.d., 5μm particle size; Agilent technologies). The column was eluted with 0.1% (v/v) trifluroacetic acid (TFA) in water and 0.1% (v/v) TFA in acetonitrile. The bound proteins were eluted with an increasing gradient of 2 to 100% acetonitrile for 40min at a flow rate of 1mL/min.
Molecular weight determination by SDS-PAGEThe lipase purified by ion exchange chromatography was subjected to SDS-PAGE (4% and10%, v/v acrylamide stacking and separating gel).11 Next, the resulting gel was stained with Coomassie Brilliant Blue, and the molecular weight was determined by comparing the protein band with a standard protein marker containing mixtures of phosphorylase β (97kDa), albumin (66kDa), ovalbumin (45kDa), carbonic anhydrase (30kDa), trypsin inhibitor (20kDa) and α-lactalbumin (14kDa).
Mass spectrometric sequence analysis of lipase from Acinetobacter sp. AU07MALDI-TOF MS is a sensitive technique used to analyse protein sequences by screening the masses of peptides and small proteins. By measuring the masses of peptides, it is possible to determine the sequence of the protein. The purified lipase was excised from the SDS-PAGE gel and subjected to proteolysis by trypsin, and the resulting peptides were analysed using MALDI-TOF. The mass of the residues of the purified lipase was obtained and used to search against the MASCOT database for peptide mass fingerprinting.
Biochemical characterization of purified Acinetobacter sp. AU07 lipaseEffect of temperature on lipase activity and stabilityThe optimum temperature for lipase activity was measured by incubating aliquots of purified enzyme with 4-nitrophenyl palmitate (4NPP) (1mM final concentration) and 20mM sodium-phosphate buffer at temperatures ranging from 30 to 80°C for 10min, and the relative activity was determined following incubation.
The thermal stability of the lipase was measured by incubating aliquots of purified enzyme at 40°C, 50°C, 60°C and 70°C for 5h, after which the residual activity was determined using the following equations:
where A0 is the initial activity, A is the residual activity in time t and kd is first-order deactivation rate coefficient:From Eq. (1), the thermal deactivation profiles were determined,12 and the respective deactivation rate (kd) estimated. From Eq. (2), the half-life (t1/2) of the biocatalyst was estimated from the previously determined kd value.
Effect of pH on lipase activity and stabilityThe optimal pH was determined by assaying the purified enzyme at different hydrogen ion concentrations using the following buffers: citric acid/sodium acetate (pH 3.0–5.0), sodium phosphate (pH 6.0–7.5), Tris–HCl (pH 7.0–9.0), glycine–NaOH (pH 9.0–10.0) and phosphate–NaOH (pH 11.0).
For the pH stability studies, the purified enzyme was incubated with the respective buffers (50mM) for 1h at 30°C, and then the residual activity was determined.
Effect of organic solvents on lipase stabilityThe purified lipase was incubated with 30% (v/v) organic solvent at 30°C with constant agitation at 150rpm for 4h. Aliquots of the suspended enzyme were taken at different intervals, and the solvent was evaporated. The enzyme activity was then measured at 30°C and pH 8.0. The stability of the lipase was calculated by comparing the residual activity with the control, which was incubated without the organic solvent.
Effect of metal ions on lipase activityVarious metal salts (ZnCl2, MgSO4·7H2O, FeCl3, CaCl2·2H2O, AgNO3, MnSO4·H2O) were added to the purified lipase at concentrations of 2mM, 5mM, and 10mM and incubated at 30°C for 30min. The relative activity of the enzyme was measured as the change in the activity of the enzyme in the presence of metal salts when compared to the enzyme activity without metal salts.
Effect of inhibitors, detergents, and oxidizing agent on lipase stabilityThe stability of the purified lipase in the presence of inhibitors, detergents and oxidizing agent was evaluated by incubating the purified lipase with the inhibitors-DTT, EDTA and PMSF at a final concentrations of 2mM, 5mM and 10mM; the detergents-Tween 80, Triton X-100, SDS at a final concentration of 0.25%, 0.5% and 1%; and an oxidizing agent-H2O2 at a final concentration of 0.5%, 1%, 1.5% and 2%. The incubated enzyme (without inhibitors, detergents and oxidizing agent) was used as a reference to calculate the relative activity.
Determination of lipase kinetics using different substratesThe hydrolytic activity of Acinetobacter sp. AU07 lipase was investigated using different substrates, such as 4-nitrophenyl butyrate (4-NPB), 4-nitrophenyl laurate (4-NPL), 4-nitrophenyl myristate (4-NPM) and 4-nitrophenyl palmitate (4-NPP), at various substrate concentrations (0.2–2mM). The Michaelis–Menten enzyme kinetic constants Km and Vmax were determined from the Lineweaver–Burk and Woolf–Hanes plots.
Results and discussionIsolation and identification of bacteria producing extracellular lipaseThe bacterial strains that were isolated from distillery waste were screened for lipase secretion by selection on tributyrin agar plates. The AU07 strain that showed a larger zone of clearance around the colony was selected for further studies. The morphological and biochemical characterization revealed that the AU07 strain is aerobic, gram negative and non-motile. The strain was confirmed as Acinetobacter sp. AU07 by 16S rDNA sequencing which is in accordance with Bergey's Manual of Determinative Biology.
Optimization of culture conditions for lipase production by response surface methodologyA maximal lipase activity of 331.16U (specific activity of 38.64U/mg) was obtained by culturing the bacterium at 30°C and pH 7.0 for 16h. The lipase production reached a maximum during the stationary phase and then gradually decreased. The fermentation time to obtain maximum lipase activity is similar to that of lipase production by Acinetobacter sp. BK44 where maximal lipase activity occurs after 12h of incubation.13
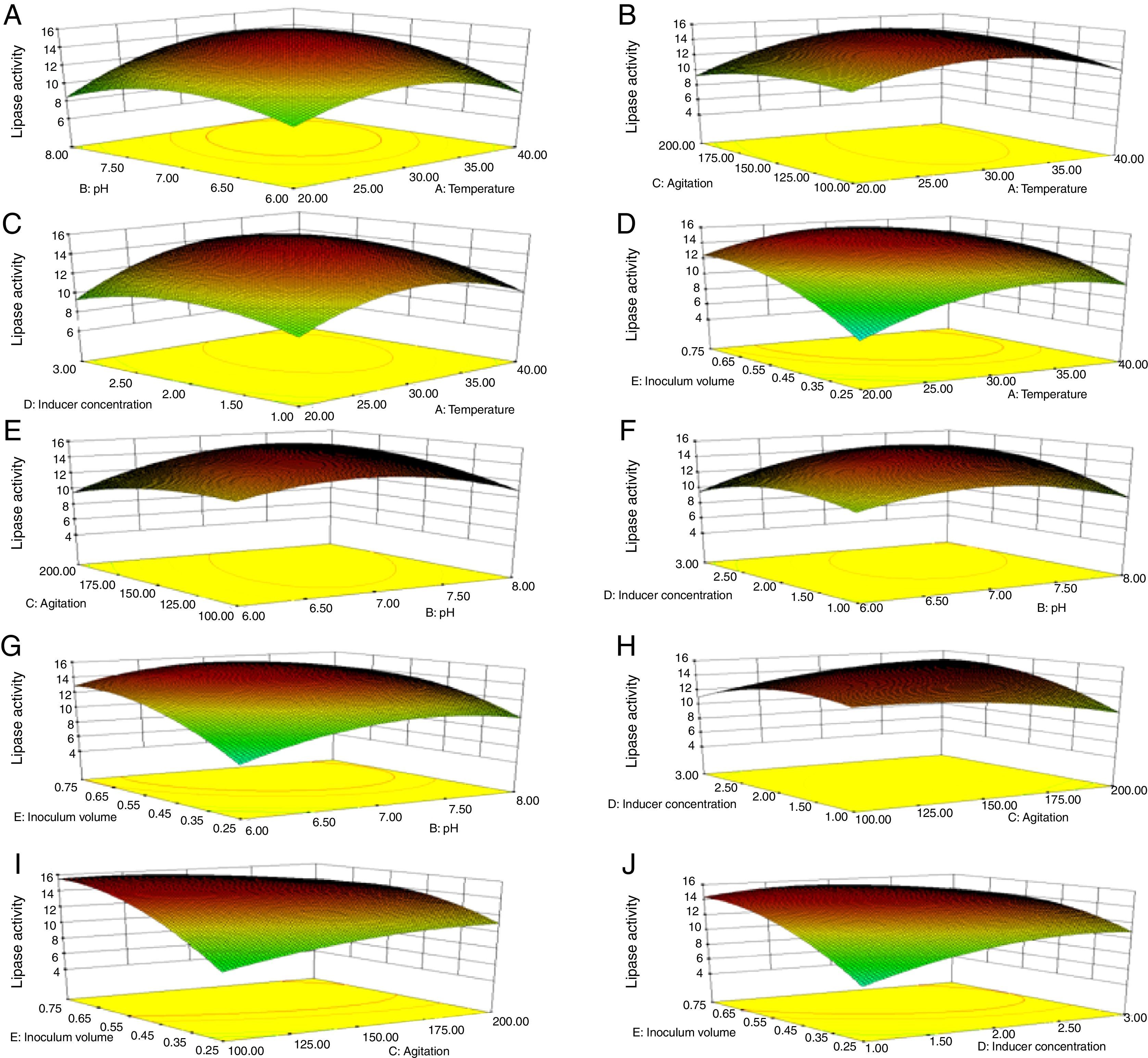
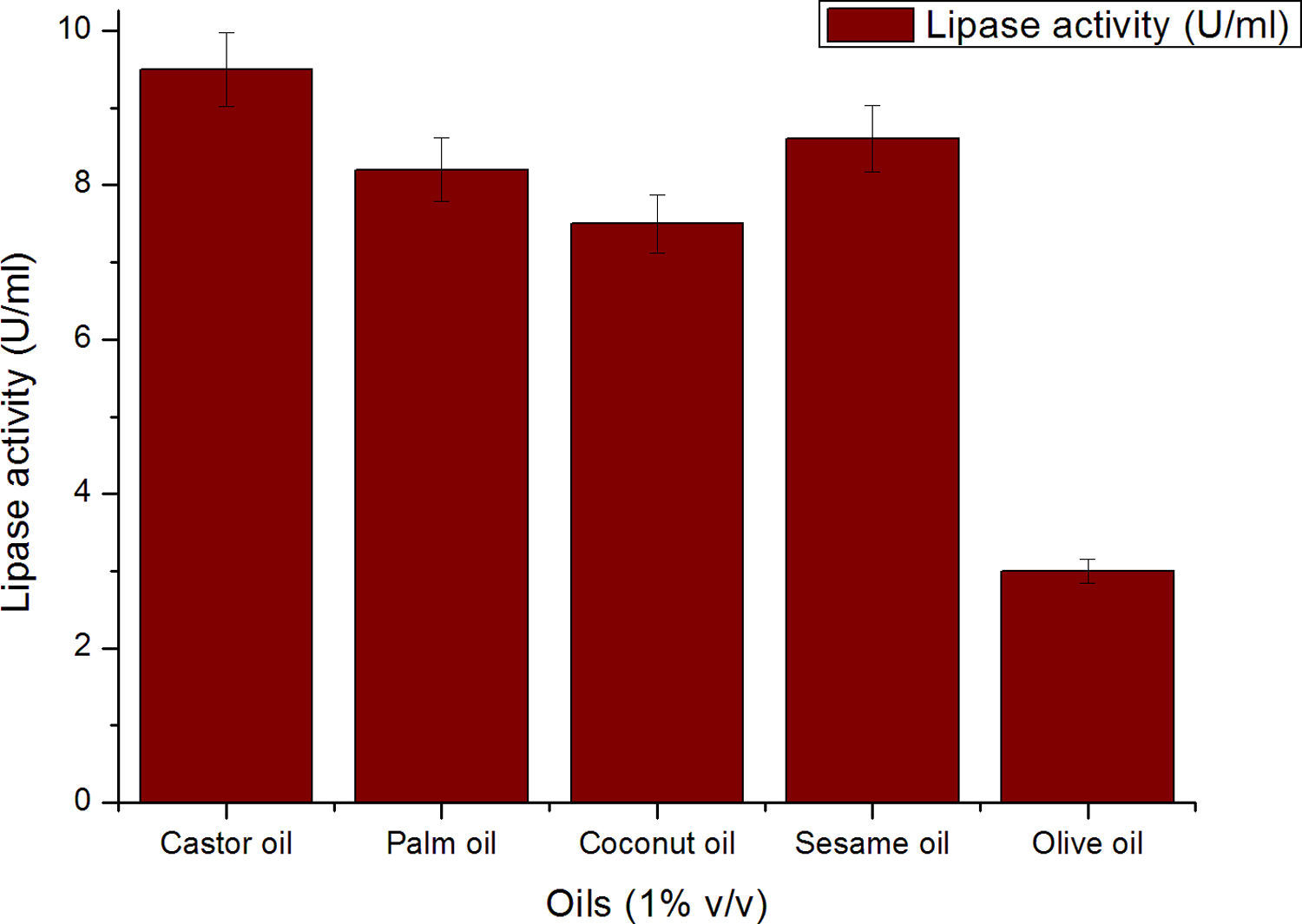
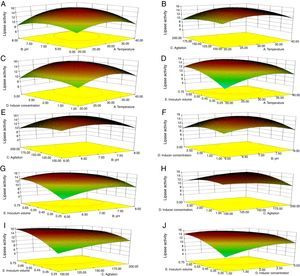
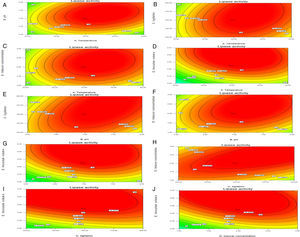
Though the lipase production from this strain is constitutive, the incorporation of oils in the medium increased the enzyme production.14–16 Earlier reports demonstrated lipase production in a medium containing lipidic substrates as the sole carbon source with an organic nitrogen source,14 but sometimes, lipase production was repressed by long chain fatty acid esters.15 Castor oil and sesame oil induced lipase production when compared with other lipid/oil sources (Supplemental Fig. S1). Castor oil is the source of ricinoleic acid which induces lipase production.17 The optimization of culture conditions one factor at a time is time consuming; therefore, response surface methodology, which is a statistical method, was used to determine the optimal conditions for lipase production.18,19 The analysis of variance for the response surface quadratic model for lipase production is shown in Table 1. The 2D contour plots and 3D response surface are graphical representations of the regression equation. They were plotted based on the model regression equation to evaluate the interactions among the tested variables and determine the optimum level of each factor to maximize lipase production (Fig. 1).
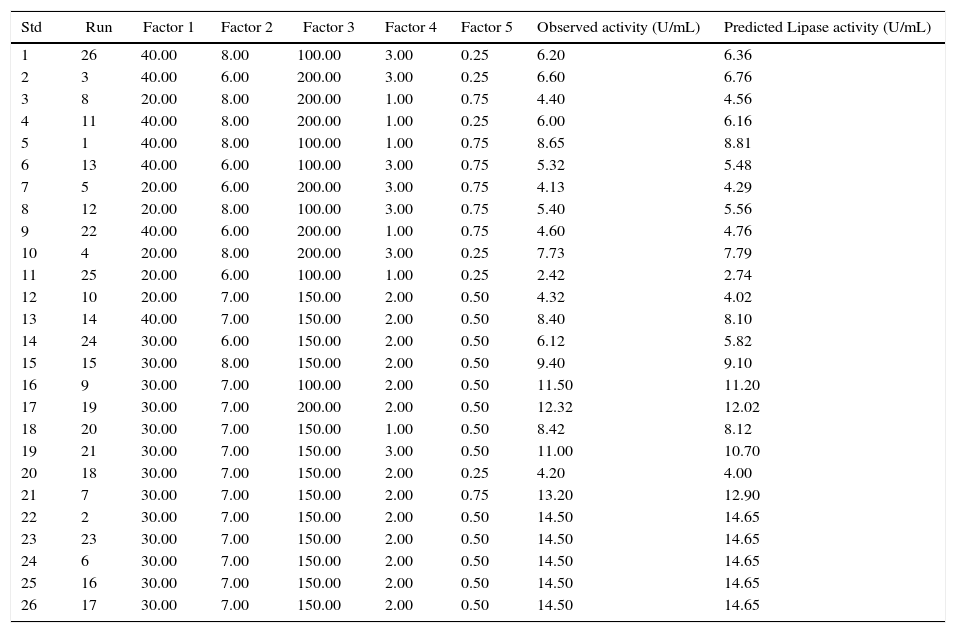
Experimental designs used in RSM studies with two independent variables showing the observed and the predicted values of lipase production.
| Std | Run | Factor 1 | Factor 2 | Factor 3 | Factor 4 | Factor 5 | Observed activity (U/mL) | Predicted Lipase activity (U/mL) |
|---|---|---|---|---|---|---|---|---|
| 1 | 26 | 40.00 | 8.00 | 100.00 | 3.00 | 0.25 | 6.20 | 6.36 |
| 2 | 3 | 40.00 | 6.00 | 200.00 | 3.00 | 0.25 | 6.60 | 6.76 |
| 3 | 8 | 20.00 | 8.00 | 200.00 | 1.00 | 0.75 | 4.40 | 4.56 |
| 4 | 11 | 40.00 | 8.00 | 200.00 | 1.00 | 0.25 | 6.00 | 6.16 |
| 5 | 1 | 40.00 | 8.00 | 100.00 | 1.00 | 0.75 | 8.65 | 8.81 |
| 6 | 13 | 40.00 | 6.00 | 100.00 | 3.00 | 0.75 | 5.32 | 5.48 |
| 7 | 5 | 20.00 | 6.00 | 200.00 | 3.00 | 0.75 | 4.13 | 4.29 |
| 8 | 12 | 20.00 | 8.00 | 100.00 | 3.00 | 0.75 | 5.40 | 5.56 |
| 9 | 22 | 40.00 | 6.00 | 200.00 | 1.00 | 0.75 | 4.60 | 4.76 |
| 10 | 4 | 20.00 | 8.00 | 200.00 | 3.00 | 0.25 | 7.73 | 7.79 |
| 11 | 25 | 20.00 | 6.00 | 100.00 | 1.00 | 0.25 | 2.42 | 2.74 |
| 12 | 10 | 20.00 | 7.00 | 150.00 | 2.00 | 0.50 | 4.32 | 4.02 |
| 13 | 14 | 40.00 | 7.00 | 150.00 | 2.00 | 0.50 | 8.40 | 8.10 |
| 14 | 24 | 30.00 | 6.00 | 150.00 | 2.00 | 0.50 | 6.12 | 5.82 |
| 15 | 15 | 30.00 | 8.00 | 150.00 | 2.00 | 0.50 | 9.40 | 9.10 |
| 16 | 9 | 30.00 | 7.00 | 100.00 | 2.00 | 0.50 | 11.50 | 11.20 |
| 17 | 19 | 30.00 | 7.00 | 200.00 | 2.00 | 0.50 | 12.32 | 12.02 |
| 18 | 20 | 30.00 | 7.00 | 150.00 | 1.00 | 0.50 | 8.42 | 8.12 |
| 19 | 21 | 30.00 | 7.00 | 150.00 | 3.00 | 0.50 | 11.00 | 10.70 |
| 20 | 18 | 30.00 | 7.00 | 150.00 | 2.00 | 0.25 | 4.20 | 4.00 |
| 21 | 7 | 30.00 | 7.00 | 150.00 | 2.00 | 0.75 | 13.20 | 12.90 |
| 22 | 2 | 30.00 | 7.00 | 150.00 | 2.00 | 0.50 | 14.50 | 14.65 |
| 23 | 23 | 30.00 | 7.00 | 150.00 | 2.00 | 0.50 | 14.50 | 14.65 |
| 24 | 6 | 30.00 | 7.00 | 150.00 | 2.00 | 0.50 | 14.50 | 14.65 |
| 25 | 16 | 30.00 | 7.00 | 150.00 | 2.00 | 0.50 | 14.50 | 14.65 |
| 26 | 17 | 30.00 | 7.00 | 150.00 | 2.00 | 0.50 | 14.50 | 14.65 |
The table includes the factors influencing lipase production. Factor 1: temperature (°C), Factor 2: pH, Factor 3: agitation (rpm), Factor 4: inducer concentration (%).
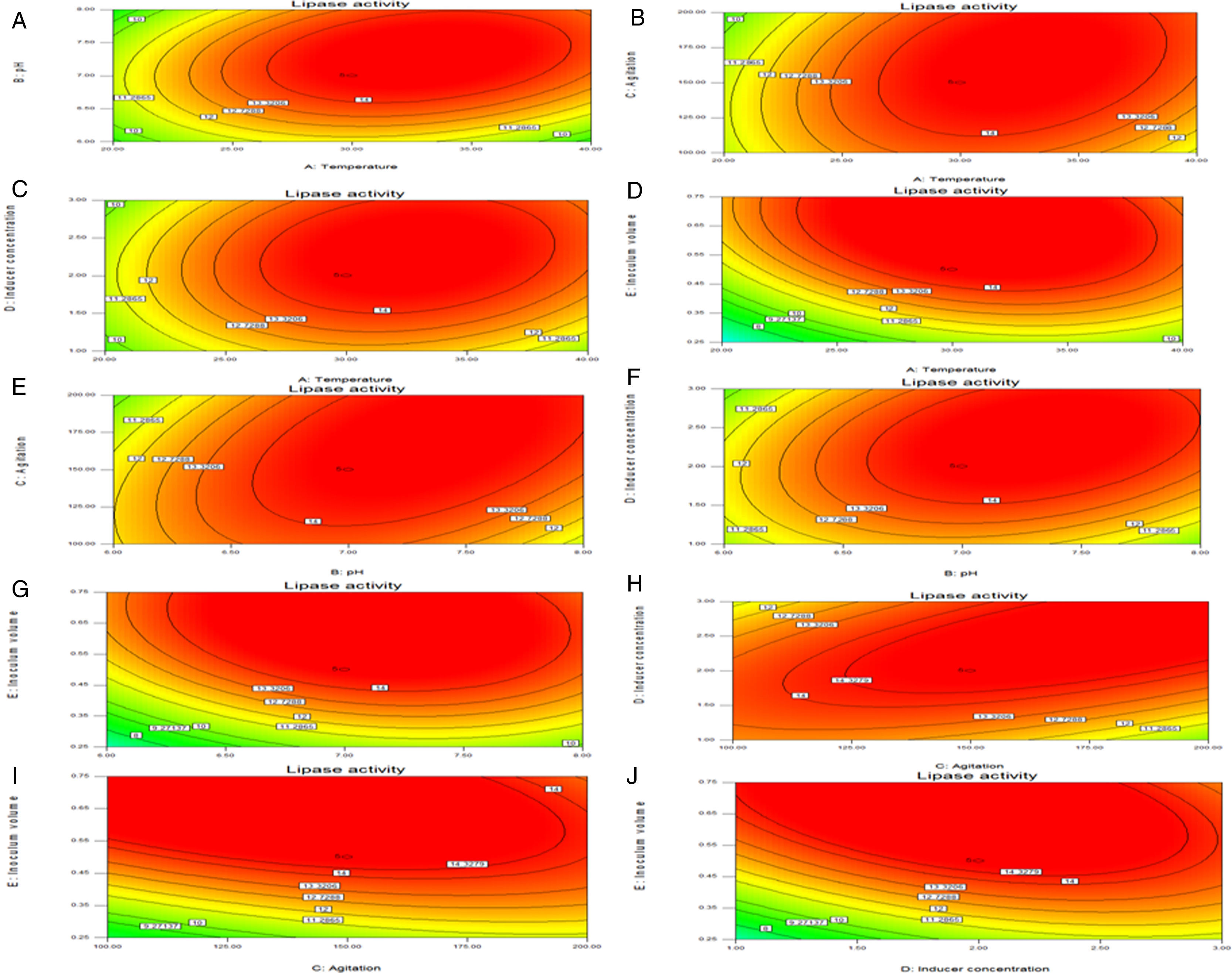
Response surface plots of the impact of various factors on optimal production of the AU07 lipase. Each figure depicts the impact of specific factors on lipase production. The variables investigated were temperature, pH, agitation, inducer concentration, and inoculum volume. (A) Temperature and pH, (B) temperature and agitation, (C) temperature and inducer, (D) temperature and inoculum volume, (E) pH and agitation, (F) pH and inducer, (G) pH and inoculum volume, (H) agitation and inducer, (I) agitation and inoculum volume, and (J) inducer and inoculum volume.
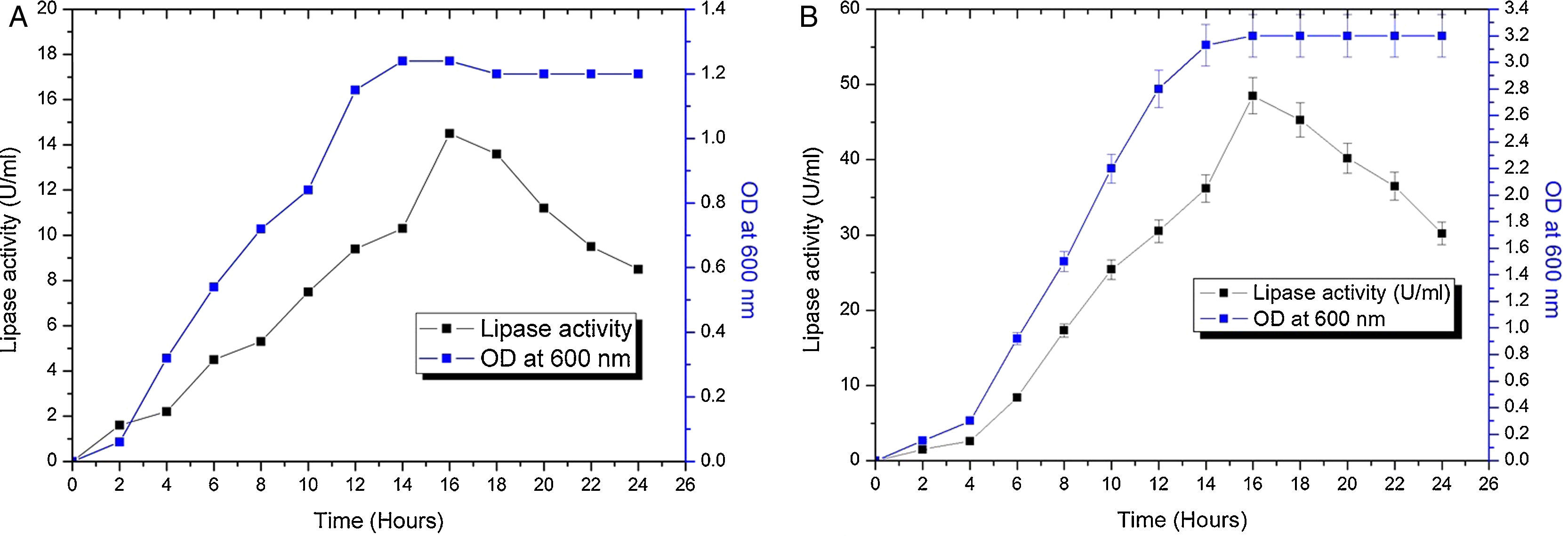
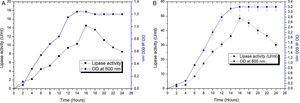
The contours were plotted considering one variable constant at its central level, while the other was varied within their experimental ranges. The data from the 2D contour plots of lipase production by Acinetobacter sp. AU07 (Supplemental Fig. S2) suggest a significant increase in lipase production resulting from all the factors investigated: 0.5% (v/v) inoculum size, 30°C fermentation temperature, pH 7, 2% (v/v) inducer concentration and agitation at 150rpm. The optimal temperature for lipase production observed in this study is similar to those of Acinetobacter radioresistens20 and Acinetobacter calcoaceticus LP009.21 Our standardized conditions gave an optimum lipase production of 14.5U/mL (Fig. 2A).
Growth curve and lipase production from Acinetobacter sp. AU07 in shake flask and bioreactor. (A) Growth curve and lipase production from Acinetobacter sp. AU07 in a shake flask (30°C, pH 7.0, 0.5%, v/v inoculum size, 2%, v/v inducer concentration, 150rpm). The lipase activity was measured using 1mM 4-NP as the substrate. The reaction was incubated at 30°C for 10min, and the absorbance was measured at 410nm. The OD was converted to lipase activity (U/mL) and the graph was plotted. Each data point is the mean of three replicates. (B) Growth curve and production of lipase from Acinetobacter sp. AU07 in a bioreactor (3L) (30°C, pH 7.0, 0.5%, v/v inoculum size, 2%, v/v inducer concentration, 150rpm, 1.5 vvm aeration). The lipase activity was measured using 4-NP (1mM) as the substrate. The reaction was incubated at 30°C for 10min and OD was measured at 410nm. The OD was converted to lipase activity (U/mL) and the graph was plotted. Each data point is the mean of three replicates.
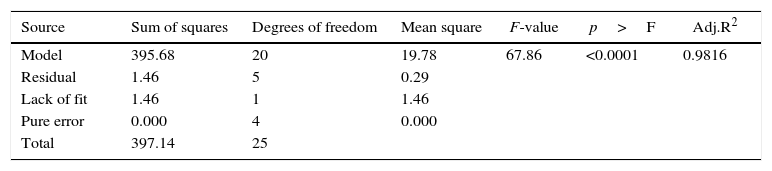
The model predicted a maximum lipase production of 16.08U/mL at 34°C, and pH 7.8, with castor oil as the inducer (2.3%, v/v), at an inoculum volume (0.44%, v/v), and with agitation at 199rpm. To validate the predicted response, the lipase production was tested experimentally under optimal culture conditions, and the maximum lipase activity was 15.84U/mL. The lipase production from this strain cultured under optimized conditions is similar to that of A. calcoaceticus.22 The significance of the response surface quadratic model was analysed by ANOVA (Table 2), and used to generate a quadratic equation for lipase activity (Y). The adjusted R2 (adj. R2=0.9816) is high and confirmed the model is significant. Additionally, the statistical coefficients confirmed that the model is significant and can be applied to increase lipase production. The production of lipase increased to 48U/mL when cultivated in a 3L bioreactor under controlled conditions (Fig. 2B).
The equation above gives the relationship between lipase activity and the tested variables, where A is the temperature, B is pH, C is agitation, D is inducer concentration, and E is inoculum volume.
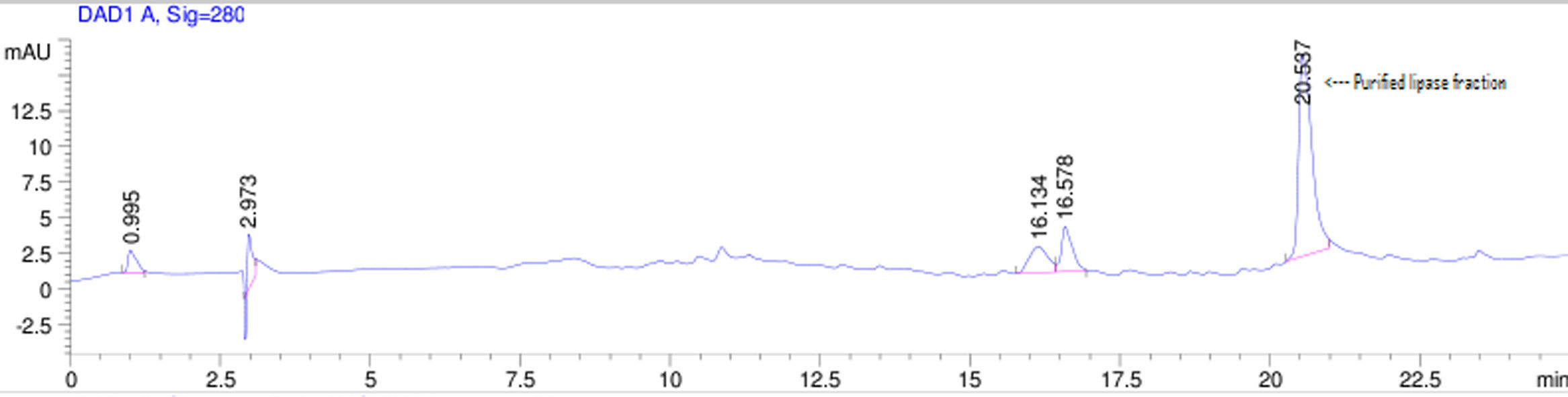
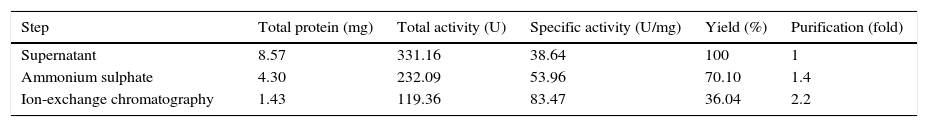
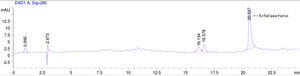
Purification of lipaseThe Acinetobacter sp. AU07 lipase (ASL) was partially purified by ammonium sulfate precipitation (30–60% saturation) and further purified to homogeneity by anion exchange chromatography. The lipase was successfully purified from cell-free crude supernatant with an overall yield of 36.04%, with a 2.2 fold purification and a specific activity of 83.47U/mg. The purification results are summarized in Table 3. The Acinetobacter sp. AU07 lipase was eluted in an increasing gradient of NaCl (0.1–1M). The maximal lipase activity was obtained in the fraction eluted with 600mM of NaCl, indicating the strong binding of the protein to the ion-exchange resin. The purity of the enzyme was analysed by RP-HPLC. The lipase was eluted as a single peak with a retention time of 20.537min as shown in the Supplemental Fig. S3.
Purification table of Acinetobacter sp. AU07 lipase.
| Step | Total protein (mg) | Total activity (U) | Specific activity (U/mg) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| Supernatant | 8.57 | 331.16 | 38.64 | 100 | 1 |
| Ammonium sulphate | 4.30 | 232.09 | 53.96 | 70.10 | 1.4 |
| Ion-exchange chromatography | 1.43 | 119.36 | 83.47 | 36.04 | 2.2 |
A two-step purification of the lipase by ammonium sulfate (60% saturation) and ion exchange chromatography (0.1–1M NaCl elution). The assay was performed by incubating the enzyme at 30°C for 10min with 1mM 4-NP as the substrate. The protein concentration was estimated by the standard Bradford assay procedure. Each value is the average of three replicates.
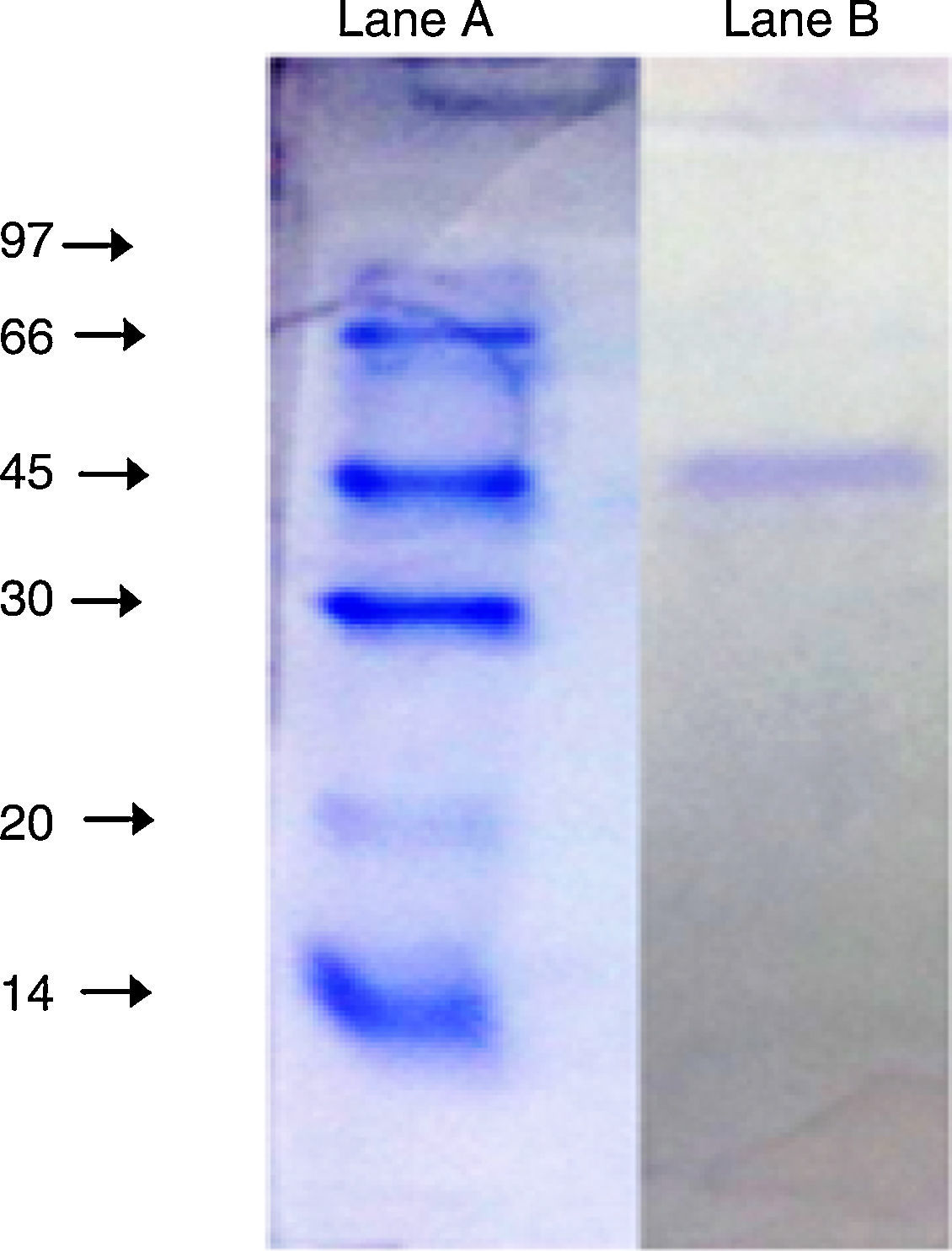
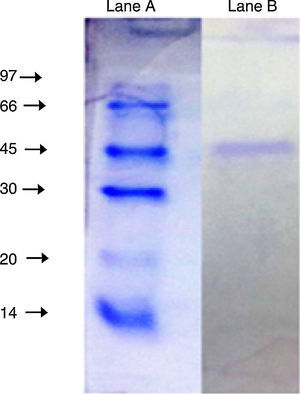
The purified enzyme resolved into a single band on an SDS-PAGE gel (Fig. 3) had an estimated molecular weight of 45kDa, which is the same as the A. radioresistens CMC-1 lipase.23
SDS-PAGE (10%) of the purified lipase from Acinetobacter sp. AU07. Lane A: Molecular mass marker proteins: phosphorylase β (97kDa), albumin (66kDa), ovalbumin (45kDa), carbonic anhydrase (30kDa), trypsin inhibitor (20kDa) and α-lactalbumin (14kDa). Lane B: Purified lipase (10μg) after ion-exchange chromatography showing single band.
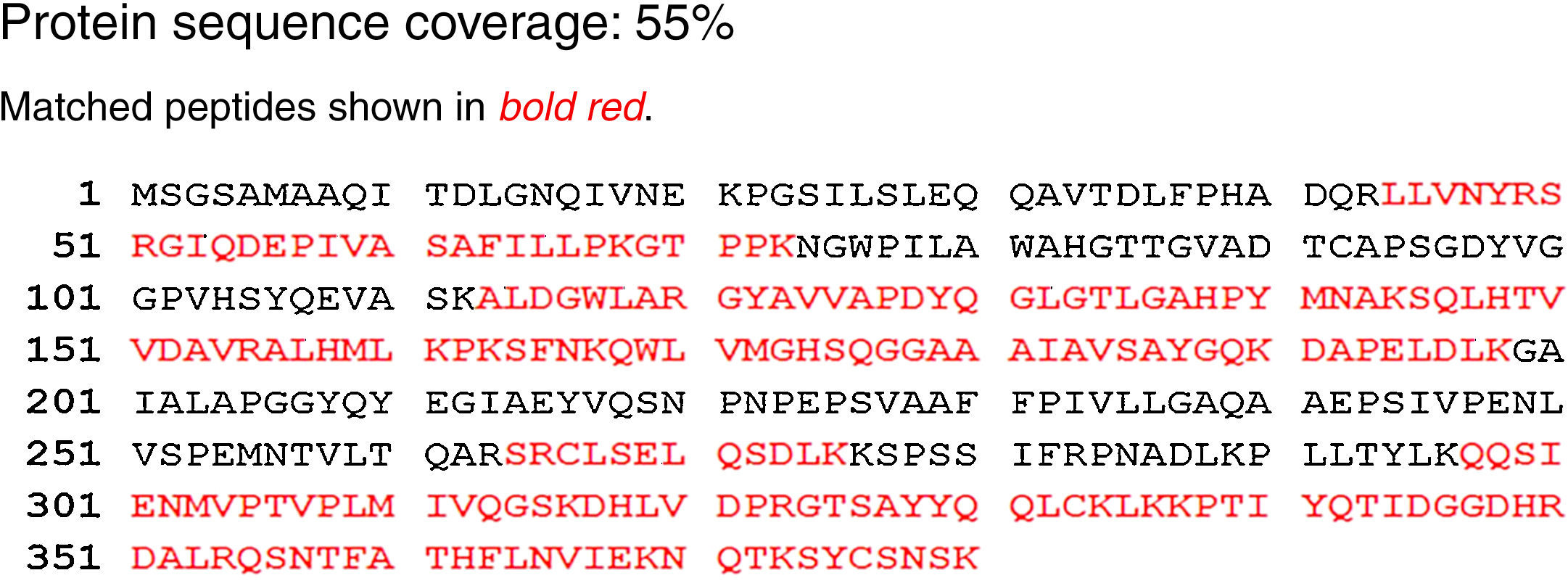
The lipase in this study has 55% homology to the lipase from A. calcoaceticus PHEA-2 followed by the lipases from Acinetobacter pitti and Acinetobacter baumanii. The partial amino acid sequence is shown in Fig. 4. The molecular mass and the isoelectric point (pI) were predicted to be 41.196kDa and 7.14, respectively. The peptide masses and the matching amino acid sequences of peptides are listed in the Supplemental Fig. S4.
The activity of a variety of lipases primarily depends on the catalytic triad typically formed by Ser, His, and Asp residues. This triad was present in the isolated Acinetobacter sp. lipase sequence (Supplemental Fig. S5). Partial amino acid sequencing confirmed that the enzyme isolated in this study possesses the characteristics of the lipase enzyme family.
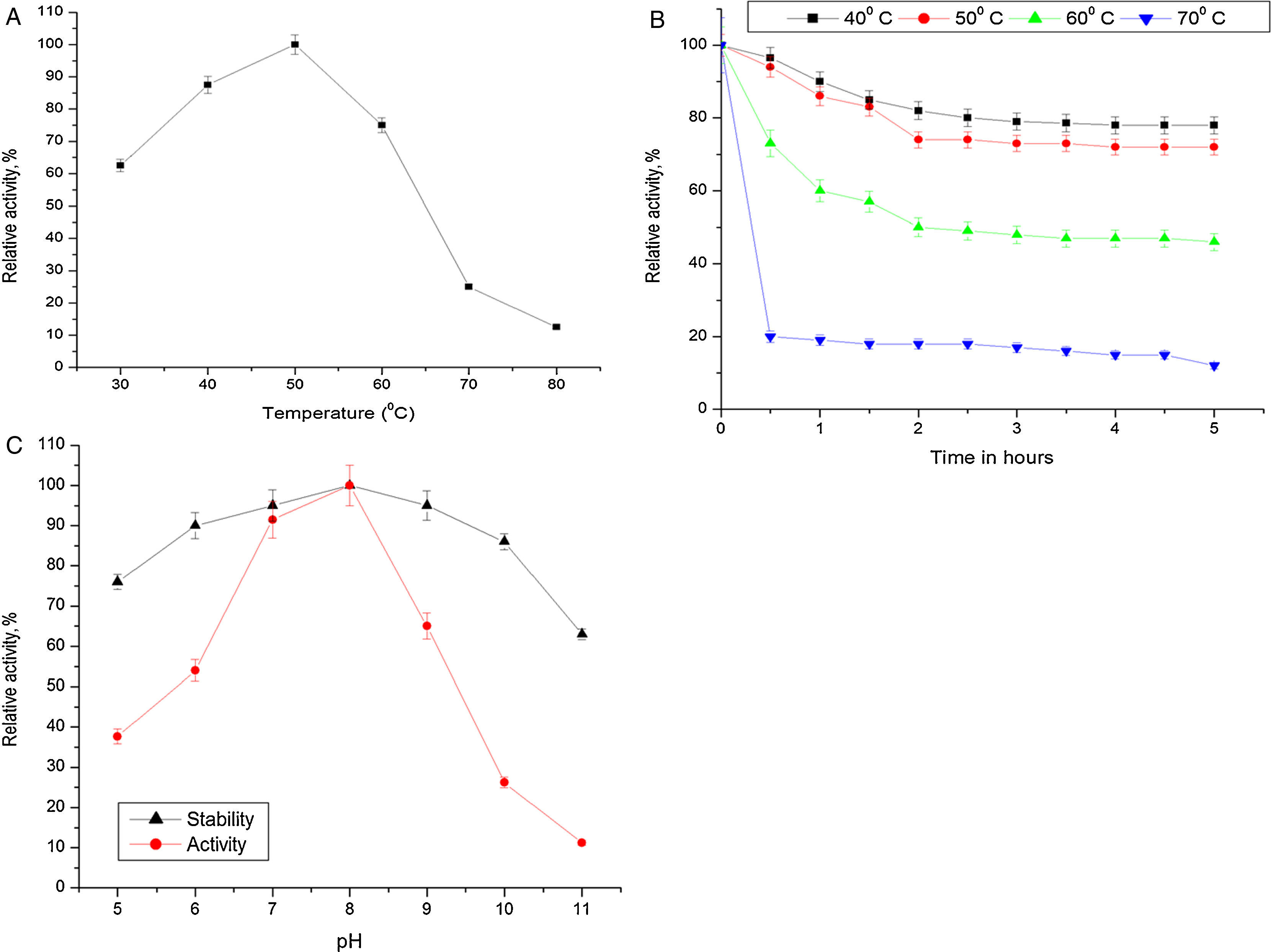
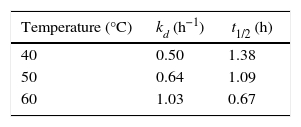
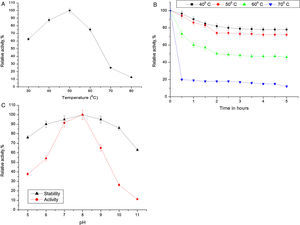
Biochemical characterization of purified Acinetobacter sp. AU07 lipaseEffect of temperature on activity and stability of lipaseThe optimum temperature for lipase activity was 50°C (Fig. 5A), whereas approximately 75% and 25% activity were observed at 60°C and 70°C, respectively. The optimum temperature for lipase activity was similar to that of lipase from A. calcoaceticus LP009.21 The stability of the lipase at different temperatures was determined by incubating the enzyme at different temperatures for 5h and assaying the enzyme at 50°C. After 5h, the enzyme retained 78% activity at 40°C and 72% activity at 50°C (Fig. 5B), which demonstrates that the enzyme is thermo stable. A linear increase in the deactivation constants (kd) was observed when the temperature was increased from 40°C to 60°C. The half-life of the enzyme was 1h 38min at 40°C, 1h 9min at 50°C and 1h and 7min at 60°C. The corresponding thermal deactivation constants (kd) and half-life (t1/2) of the purified lipase are shown in Table 4.
(A) Effect of temperature on the activity of Acinetobacter sp. AU07. The reaction mixture was incubated at different temperatures (30–80°C), and the enzyme activity was assayed under standard assay conditions. (B) Effect of temperature on the stability of the Acinetobacter sp. AU07 lipase. To determine the thermal stability, the enzyme was pre-incubated at various temperatures (40–70°C) for 5h, and the enzyme activity was assayed under standard assay conditions. (C) Effect of pH on the activity and stability of the Acinetobacter sp. AU07 lipase. For the enzyme activity assays, the reaction was assayed at various pH levels under standard assay conditions. For the enzyme stability assays, the enzyme was pre-incubated with various pH buffers (50mM) of pH (5–11) at 30°C for 1h, and then the residual activity was determined by assaying under standard assay conditions.
Half-life and thermal deactivation constants at different temperatures.
| Temperature (°C) | kd (h−1) | t1/2 (h) |
|---|---|---|
| 40 | 0.50 | 1.38 |
| 50 | 0.64 | 1.09 |
| 60 | 1.03 | 0.67 |
The half-life and thermal deactivation constants were determined by incubating aliquots of pure enzyme at various temperatures for 5h, after which, the residual enzyme activity was determined.
Most of the lipases from Acinetobacter sp. are alkali stable, 24,25 losing their activity at low pH. This could be due to the loss of the coordinating metal ion Ca2+ metal ion from the active site at low pH.26 The purified lipase in this study showed the highest activity at pH 8.0 which is similar to the Acinetobacter sp. CR9 lipase 27 and the Acinetobacter baylyi lipase.28 The A. radioresistens CMC-1 lipase had a pH optima at 10.5,23 whereas, A. calacoaceticus 1–7 had a pH optima of 9.29 In contrast, the lipases from A. calcoaceticus LP009 and Acinetobacter sp.13,21 had pH optima of 7 and 6 respectively. ASL presented a relative activity of 65% at pH 9, 26% at pH 10, and only 11% at pH 11. The activity was not reduced significantly after incubation for 1h at 30°C (Fig. 5C).
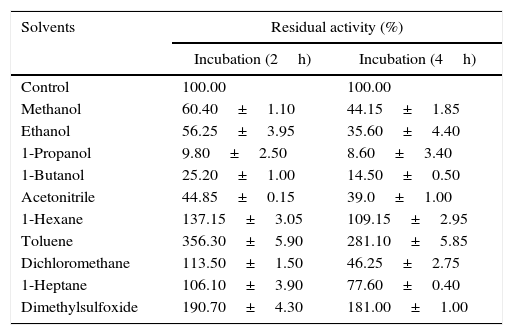
Effect of organic solvents on lipase stabilityThe stripping of water from an enzyme primarily occurs in the presence of polar solvents and to a lesser degree in the presence of hydrophobic solvents determined by logp values. For instance, methanol desorbed approximately 60% of the water bound to the enzyme, while hexane only desorbed 0.5%.30 Solvents with high logp values cause less inactivation of enzyme than solvents with low logp values.31 Many researchers have previously reported the solvent stable nature of lipases,24,32–34 thus the stability of Acinetobacter sp. AU07 lipase in different solvents was studied (Table 5). The polar solvents (low logp) reduced the enzyme activity, whereas the non-polar solvents (higher logp) increased the enzyme activity. Among the polar solvents, DMSO increased the activity 1.9 fold. Toluene a non-polar solvent, increased the activity 3.5-fold when incubated for 2h and 2.8-fold when incubated for 4h. When incubated for 4h each at 30°C, other non-polar solvents also increased the activity considerably, viz., n-hexane 1-fold, dichloromethane 0.4-fold, and n-heptane 0.7-fold.
Effect of organic solvents on the Acinetobacter sp. AU07 lipase.
| Solvents | Residual activity (%) | |
|---|---|---|
| Incubation (2h) | Incubation (4h) | |
| Control | 100.00 | 100.00 |
| Methanol | 60.40±1.10 | 44.15±1.85 |
| Ethanol | 56.25±3.95 | 35.60±4.40 |
| 1-Propanol | 9.80±2.50 | 8.60±3.40 |
| 1-Butanol | 25.20±1.00 | 14.50±0.50 |
| Acetonitrile | 44.85±0.15 | 39.0±1.00 |
| 1-Hexane | 137.15±3.05 | 109.15±2.95 |
| Toluene | 356.30±5.90 | 281.10±5.85 |
| Dichloromethane | 113.50±1.50 | 46.25±2.75 |
| 1-Heptane | 106.10±3.90 | 77.60±0.40 |
| Dimethylsulfoxide | 190.70±4.30 | 181.00±1.00 |
The enzyme was pre-incubated for 2 and 5h with different organic solvents (30%, v/v) while shaking (150rpm) at 30°C and assayed under standard assay conditions. The stability of the lipase was calculated by comparing the residual activity with that of the control, which was incubated without the organic solvent and whose enzyme activity was taken as 100%. The standard deviation for all the values is <±5.0%.
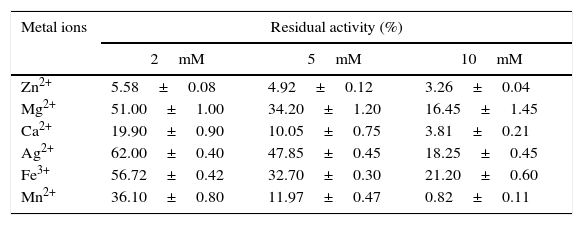
Metal ions decreased the activity of the enzyme. The lipase retained only 3% of its initial activity with a 10mM Zn2+ concentration (Table 6). Several authors observed similar results using Zn2+, 23,35–39 although the inhibitory effect is not yet known. The decrease in the activity of the enzyme in the presence of metals ions implies that the AU07 lipase does not require metal ions for its activity.
Effect of metal ions on the activity of the Acinetobacter sp. AU07 lipase.
| Metal ions | Residual activity (%) | ||
|---|---|---|---|
| 2mM | 5mM | 10mM | |
| Zn2+ | 5.58±0.08 | 4.92±0.12 | 3.26±0.04 |
| Mg2+ | 51.00±1.00 | 34.20±1.20 | 16.45±1.45 |
| Ca2+ | 19.90±0.90 | 10.05±0.75 | 3.81±0.21 |
| Ag2+ | 62.00±0.40 | 47.85±0.45 | 18.25±0.45 |
| Fe3+ | 56.72±0.42 | 32.70±0.30 | 21.20±0.60 |
| Mn2+ | 36.10±0.80 | 11.97±0.47 | 0.82±0.11 |
The pure enzyme was incubated with metal salts of different concentrations, and the residual enzyme activity was determined under standard assay conditions. The enzyme activity without metal salts was taken as 100%. Each value presented here is an average of three replicates.
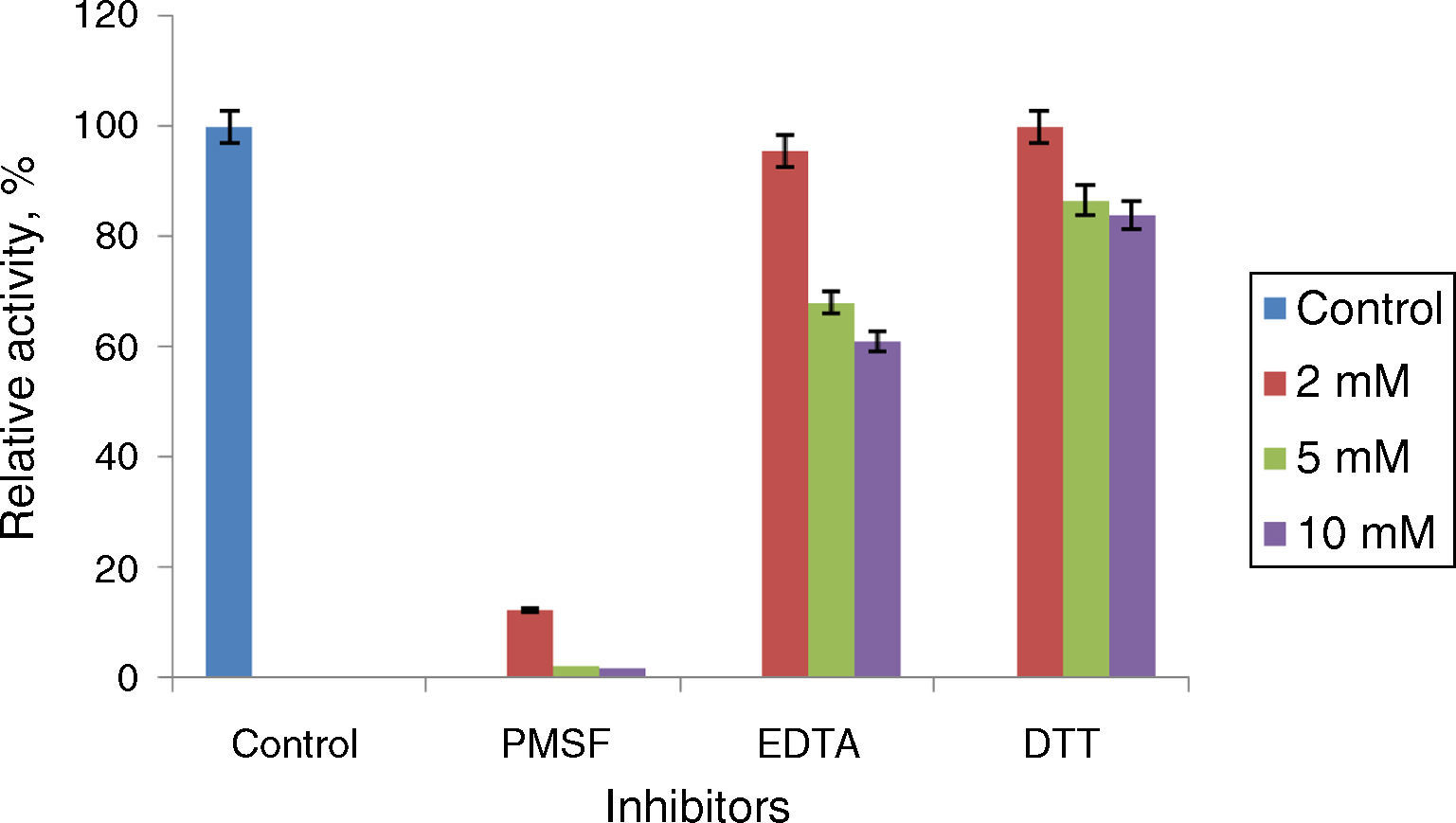
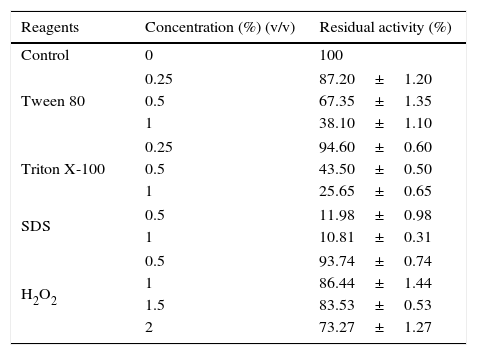
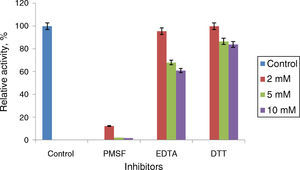
Phenyl methylsulphonyl fluoride (PMSF: an inhibitor of serine lipase) inhibited the lipase activity (88% inhibition at 2mM concentration). Similar results were observed in the lipases from A. radioresistens CMC-123 and Bacillus stearothermophilus P140 suggesting the involvement of a serine residue in the active site. There was minimal inhibition by EDTA and DTT (Fig. 6). The decrease in the enzyme activity in the presence of metal ions and the absence of inhibition of the enzyme activity by EDTA indicate that metal ions are not required for the lipase activity. These results suggest that this lipase may be a member of serine-enzyme family. The effect of detergents such as SDS, Tween 80 and Triton X-100 on the enzyme activity were studied. SDS inhibited the enzyme activity; however, there was minimal loss in enzyme activity when incubated with 0.25mM Tween 80 or Triton X-100 (Table 7). Oxidizing agents (hydrogen peroxide) had minimal effect on the enzyme activity, and the enzyme retained 94% (at 0.5mM) and 73% (at 2mM) of its enzyme activity, respectively.
Effect of inhibitors on Acinetobacter sp. AU07 lipase activity. The enzyme was incubated with different inhibitors at 30°C for 1h, and the enzyme activity was assayed under standard assay procedure conditions. The enzyme incubated without inhibitors was used as a reference to calculate the relative activity. Each value presented here is an average of three replicates.
Effect of detergents and oxidizing agent on the activity of the Acinetobacter sp. AU07 lipase.
| Reagents | Concentration (%) (v/v) | Residual activity (%) |
|---|---|---|
| Control | 0 | 100 |
| Tween 80 | 0.25 | 87.20±1.20 |
| 0.5 | 67.35±1.35 | |
| 1 | 38.10±1.10 | |
| Triton X-100 | 0.25 | 94.60±0.60 |
| 0.5 | 43.50±0.50 | |
| 1 | 25.65±0.65 | |
| SDS | 0.5 | 11.98±0.98 |
| 1 | 10.81±0.31 | |
| H2O2 | 0.5 | 93.74±0.74 |
| 1 | 86.44±1.44 | |
| 1.5 | 83.53±0.53 | |
| 2 | 73.27±1.27 | |
The enzyme was incubated with different detergents and oxidizing agents of various concentrations at 30°C for 1h and then assayed under standard assay conditions. The enzyme incubated without detergents and oxidizing agents was used as a reference and was used to calculate the relative enzyme activity. Each value presented here is an average of three replicates.
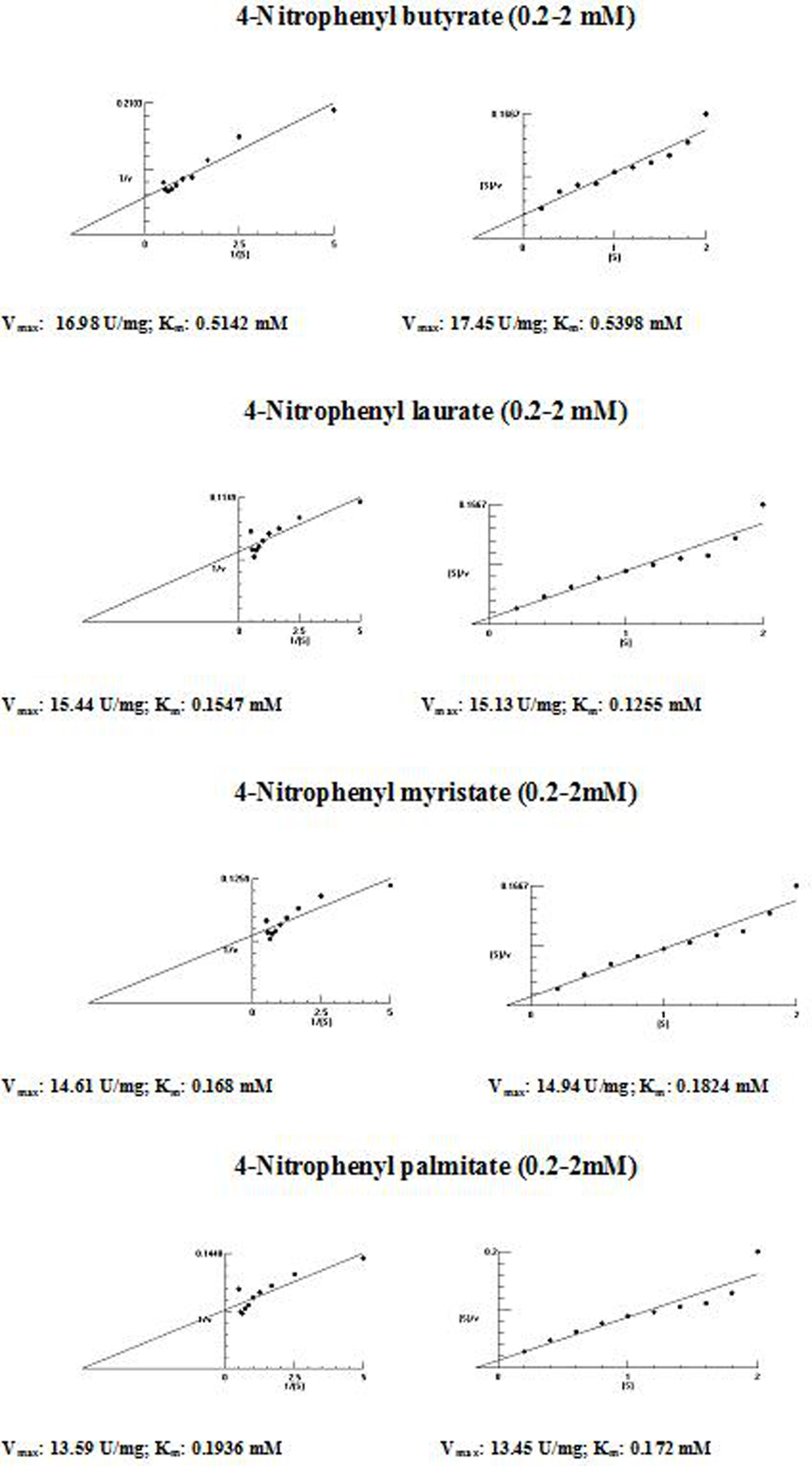
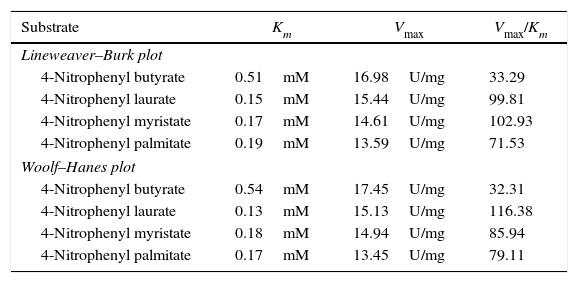
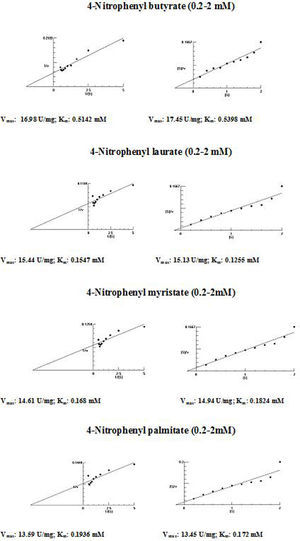
The kinetic parameters (Km and Vmax) of hydrolysis of different fatty acid esters were determined. The Km was higher for 4-nitrophenyl butyrate (short fatty acid esters), and low Km values were obtained for 4-nitrophenyl laurate, 4-nitrophenyl myristate and 4-nitrophenyl palmitate (moderate chain length fatty acid esters) indicating that the Acinetobacter sp. AU07 lipase is more specific to soluble fatty acid esters of moderate chain lengths. These results are consistent with Acinetobacter baumannii BD5 lipase.41 A low Km value indicates the enzyme has a high affinity for a substrate, whereas a high Km value indicates a low affinity. The comparison of kinetic constants (Km and Vmax) obtained for various substrates (4-nitrophenyl esters) suggest that the hydrolytic activity of this lipase was specific to moderate chain fatty acid esters. This lipase was found to have a lower Km value for 4-Nitrophenyl laurate when compared with the other substrates. This suggests that the lipase has a higher affinity towards 4-nitrophenyl laurate.
The Vmax values for 4-NPL, 4-NPM and 4-NPP were high and were similar in range (Table 8). The Vmax/Km ratio was different for all substrates used, but for moderate and long chain fatty acid esters, it exceeded the ratio for p-NPB. The Km and Vmax values determined by the Lineweaver–Burk and Woolf–Hanes plots for the different substrates are shown in Supplemental Fig. S6.
Km and Vmax values of the enzyme determined by Lineweaver–Burk and Woolf–Hanes plots for different substrates.
| Substrate | Km | Vmax | Vmax/Km |
|---|---|---|---|
| Lineweaver–Burk plot | |||
| 4-Nitrophenyl butyrate | 0.51mM | 16.98U/mg | 33.29 |
| 4-Nitrophenyl laurate | 0.15mM | 15.44U/mg | 99.81 |
| 4-Nitrophenyl myristate | 0.17mM | 14.61U/mg | 102.93 |
| 4-Nitrophenyl palmitate | 0.19mM | 13.59U/mg | 71.53 |
| Woolf–Hanes plot | |||
| 4-Nitrophenyl butyrate | 0.54mM | 17.45U/mg | 32.31 |
| 4-Nitrophenyl laurate | 0.13mM | 15.13U/mg | 116.38 |
| 4-Nitrophenyl myristate | 0.18mM | 14.94U/mg | 85.94 |
| 4-Nitrophenyl palmitate | 0.17mM | 13.45U/mg | 79.11 |
A thermostable organic solvent tolerant lipase from Acinetobacter sp. AU07 has been purified and characterized biochemically. The production was improved by optimizing the culturing conditions using response surface methodology. The purified enzyme showed several important properties such as activity in alkaline pH, resistance to surfactants and oxidizing agents that would prove useful in industrial applications and in the detergent and pharmaceutical industries. The biofilms of the lipase secreting Acinetobacter sp. AU07 may be used for bioremediation to treat waste water contaminated with oils. The thermostable and organic solvent tolerant nature of the lipase can be used to perform synthetic reactions in the presence of organic solvents and at high temperatures.
Conflicts of interestThe authors declare no conflicts of interest.
Special thanks to the Department of Biotechnology, Government of India, for funding Senior Research fellowship (to P. Gururaj) and programme support (BT/01/COE/07/01) to Gautam Pennathur, S. Ramalingam, and G. Nandhinidevi.