Antarctica harbors a great diversity of microorganisms, including bacteria, archaea, microalgae and yeasts. The Pseudomonas genus is one of the most diverse and successful bacterial groups described to date, but only eight species isolated from Antarctica have been characterized. Here, we present three potentially novel species isolated on King George Island. The most abundant isolates from four different environments, were genotypically and phenotypically characterized. Multilocus sequence analysis and 16S rRNA gene analysis of a sequence concatenate for six genes (16S, aroE, glnS, gyrB, ileS and rpoD), determined one of the isolates to be a new Pseudomonas mandelii strain, while the other three are good candidates for new Pseudomonas species. Additionally, genotype analyses showed the three candidates to be part of a new subgroup within the Pseudomonas fluorescens complex, together with the Antarctic species Pseudomonas antarctica and Pseudomonas extremaustralis. We propose terming this new subgroup P. antarctica. Likewise, phenotypic analyses using API 20 NE and BIOLOG® corroborated the genotyping results, confirming that all presented isolates form part of the P. fluorescens complex. Pseudomonas genus research on the Antarctic continent is in its infancy. To understand these microorganisms’ role in this extreme environment, the characterization and description of new species is vital.
Antarctica is arguably one of the planet's harshest environments.1 Intense cold, low humidity, a limited availability of liquid water, as well as periods of high solar radiation and extended total darkness, mean that the continent represents a relatively inhospitable environment for the development of the life.1 Nevertheless, Antarctica is now considered to host a very high diversity of microorganisms, including bacteria, archaea, yeasts and microalgae.2,3 As a result of intense selective pressures throughout their evolutionary history, all of these microorganisms are fully adapted to the Antarctic conditions.4,5 These adaptations are of great interest to the biotechnology industry, since they harbor the possibility of obtaining bio-products that could be used in various processes of conservation at low temperatures.6 There is therefore an increasing need to investigate the diversity and to exploit the potential of the microorganisms of the cold continent.
A bacterial genus that has appeared frequently in Antarctic research is Pseudomonas. One of the most well-studied bacterial genera, it is present in most environments and can be considered one of the most successful bacterial groups on Earth.7Pseudomonas’ success is mainly based on its low nutritional requirements and great metabolic diversity, which allows it to use numerous organic compounds as a source of both carbon and energy.8,9 In addition, these bacteria produce a large amount of secondary metabolites, which are essential for their survival and even enable some species to degrade aromatic and halogenated compounds.10,11 Based on these characteristics, the bacteria of the genus Pseudomonas represent an interesting alternative for production of different and biotechnologically useful molecules.
Despite being extremely well-studied on a global level, little is known about the species diversity of the genus Pseudomonas in Antarctica. To date, only eight species have been described on this continent: Pseudomonas antarctica, Pseudomonas meridiana, Pseudomonas proteolytica,12Pseudomonas guineae,13Pseudomonas extremaustralis,14Pseudomonas deceptionensis,15Pseudomonas prosekii16 and, most recently, Pseudomonas gregormendelii.17 In order to further our understanding of this genus’ biogeographic distribution, as well as its function as part of the ecosystem of the extreme Antarctic environment, it is of utmost importance to identify and describe new Antarctic species of Pseudomonas. With this in mind, the present study aims to carry out a phylogenetic and phenotypic characterization of four bacterial isolates originating from different environments on King George Island, Antarctica. Specifically, we set the following specific objectives: (1) an evaluation of the growth rate of different strains at different temperatures; (2) the amplification and sequencing of the 16S rRNA gene; (3) the amplification and sequencing of the genes aroE, glnS, gyrB, ileS and rpoD, all of which were analyzed both individually and, after sequence concatenation, by Multilocus Sequence Analysis (MLSA), a robust, high-resolution technique for the identification of novel species and the confirmation of species identity of new isolates18–20; and (4) a phenotypic characterization using API 20 NE and BIOLOG GN2. Based on the both the phylogenetic analysis of 16S-rRNA gene and the MLSA results, as well as on the phenotypic characterization, we show that the four isolates belong to the genus Pseudomonas and specifically to the P. fluorescens complex, and that three of them might represent novel species, while one strain could be classified within the species P. mandelii.
Materials and methodsStrains and culture conditionsDifferent Antarctic environments on King George island, South Shetland archipelago, were sampled in order to isolate specifically bacteria members of the Pseudomonas genus. These samples were collected on Austral summer season on January 2011. Samples of seawater from the Fildes bay at 5m depth (62°11′50.3″ S; 58°54′50.3″ W); freshwater from a summer lake on Fildes peninsula (62°10′9.2″ S; 58°55′29″ W); marine sediments from Fildes bay (62°13′28.8″ S; 58°58′42.7″ W) and soil from the Fildes Peninsula (62°10′9.2″ S; 58°55′29″ W); were taken in order to isolate bacteria from all environmental diversity. 10L of water samples were first filtered by a 0.1mm filters and then the samples were concentrated in a 0.2μm filter. This filter was then resuspended in 3mL of saline buffer (0.85% NaCl). 5g of sediments and soil samples were resuspended in 5mL of saline buffer solution (0.85% NaCl). 100μL of the solution were plated onto Nutrient Agar (BD™ DIFCO™, New Jersey, NJ, USA) and incubated at 4, 10, 15 and 25°C for 72h. In order to isolate bacteria of the genus Pseudomonas, all colonies were replated on Pseudomonas Isolation Agar (PIA) (SIGMA–Aldrich, Los Ángeles, CA, USA) and incubated at 4, 10, 15 and 25°C for 72h. All obtained colonies were 16S rRNA gene sequenced. The most abundant isolate of each environment was selected and designed as a particular strain. Growth rates of all selected strains were estimated in Lysogeny Broth (LB) (BD™ DIFCO™, New Jersey, NJ, USA) at 4, 10, 15, 25, 30 and 37°C. All tests were performed in 100mL flasks in 20mL medium and at constant agitation rate of 240rpm, in duplicates and on three different days (making for a total of six samples). The strain P. fluorescens A506 (ATCC® 31948™) served as a control for the phenotypic characterization.
Amplification of the 16S rRNA gene and phylogenetic analysisGenomic DNA was extracted from all strains using the GeneJET Genomic DNA Purification Kit (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer's instructions. The 16S rRNA gene was amplified by PCR using universal primers 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′). All PCR products were sequenced by Sanger method for five times (Forward and reverse). The obtained sequences were deposited in GenBank (NCBI) (Table S1). The sequences were then BLASTed 21 against the 16S rRNA GenBank database (NCBI). Sixty-one 16S rRNA gene sequences of the genus Pseudomonas were selected for the phylogenetic analysis. The 16S rRNA gene of Escherichia coli strain MG1655 K12 was used as an outgroup. Sequences of all members of the genus Pseudomonas were obtained from the database on the website www.pseudomonas.com22 and aligned using ClustalW (using standard parameters) as implemented in the software MEGA 7.0.14.23 Gaps were treated with the partial deletion method. A maximum likelihood phylogenetic tree was constructed using the Tamura-Nei model. Bootstrap values were calculated based on 1000 replicas.
MLSAMLSA was performed on six housekeeping genes: 16S-rRNA, aroE (Shikimato 5-dehydrogenase), glnS (glutaminyl-tRNA transferase), gyrB (DNA gyrase subunit B), ileS (isoleucine-tRNA transferase), and rpoD (RNA polymerase sigma factor). PCR primers were taken from Andreani et al.24 For all genes, PCR consisted of an initial denaturalization step of 10min at 95°C, followed by 30 cycles of 45s at 95°C; 45s at 55°C and 1min at 72°C, and a final extension step of 10min at 72°C. All PCR products were sequenced by Sanger method for five times (Forward and reverse). The partial sequences obtained for the genes aroE, glnS, gyrB, ileS and rpoD of strains IB20, 12B3 and 6A1 were deposited in GenBank (NCBI) (Table S1). The corresponding sequences for strain KG01 were taken from its whole genome sequence, which we have published previously.25 The obtained sequences were individually BLASTed21 against GenBank in order to identify the homologues with greatest sequence identity. For the phylogenetic analysis, we concatenated the partial gene sequences after the multisequence alignment in the following order: 16S-aroE-glnS-gyrB-ileS-rpoD, resulting in a single sequence. For the same genes, we took the orthologues from 51 of the species that are most representative of the genus Pseudomonas, whose sequences were obtained from the database on the website www.pseudomonas.com.22 As an outgroup, we used the concatenate of the orthologues from the strain E. coli K12 MG1655. The phylogenetic analysis was carried out in the same way as for the 16S rRNA gene.
Phenotypic characterizationThe phenotypic characterization of the Antarctic isolates IB20, 12B3, 6A1 and KG01, as well as of the control strain P. fluorescens A506 (ATCC®31948™), was carried out using API 20 NE (Biomerieux, L’Etoile, France) and BIOLOG GN2 (BIOLOG®, Hayward, CA, USA). For both API 20 NE and BIOLOG assays, we used previously isolated colonies that had been grown for 24h at 25°C on LB agar plates (BD™ DIFCO™, New Jersey, NJ, USA). These colonies were re-suspended in saline solution (0.85% NaCl) and inoculated according to the manufacturer's instructions. Oxidase production was determined using Oxidase Reagent Droppers (Becton, Dickinson and Company, New Jersey, NJ, USA) following the manufacturer's specifications. All phenotypic experiments were performed in triplicate using six colonies in all rounds.
ResultsSelection of most abundant Pseudomonas isolates based on 16S rRNA sequenceAt the first-round selection in nutrient agar, more than 6000 colonies were obtained in all temperatures used. All colonies were replated in PIA medium in order to select only members of the Pseudomonas genus. Only 268 colonies grow in PIA in all temperatures for all environments selected. These colonies were 16S rRNA gene sequenced and the most abundant isolates for environment were used in this work. Isolate from seawater was named IB20; from freshwater 12B3; from marine sediments 6A1 and from soil KG01.
The Antarctic bacterial isolates are psychrotolerant, with optimum growth at 25°CIn order to determine the optimum growth conditions for the Antarctic isolates, we performed growth experiments at different temperatures. Growth curve analysis and the calculation of doubling times clearly showed that the optimum growth temperature is 25°C for all of the Antarctic isolates (Table 1). While no growth could be detected at 37°C, all isolates grew well at 4°C, as is typical of psychrotolerant bacteria.26 The optimum growth temperature of 25°C is shared by Pseudomonas strains across latitudes. On the Antarctic South Shetland Islands, however, this temperature has not been reached for the past 25,000 years,27 suggesting that members of the genus Pseudomonas might have colonized Antarctica before it became a polar continent.
The optimum growth temperature of the Antarctic bacterial isolates is 25°C. The table shows the doubling times of the four Antarctic bacterial strains at different temperatures (N/G=no growth).
| Doubling time (min)+SD | ||||||
|---|---|---|---|---|---|---|
| 4°C | 10°C | 15°C | 25°C | 30°C | 37°C | |
| IB20 | 341±38 | 118±21 | 65±6 | 43±8 | 75±11 | N/G |
| 12B3 | 412±46 | 132±16 | 77±4 | 56±6 | 89±11 | N/G |
| 6A1 | 381±26 | 158±18 | 69±8 | 49±9 | 78±8 | N/G |
| KG01 | 376±31 | 141±22 | 72±10 | 48±5 | 92±6 | N/G |
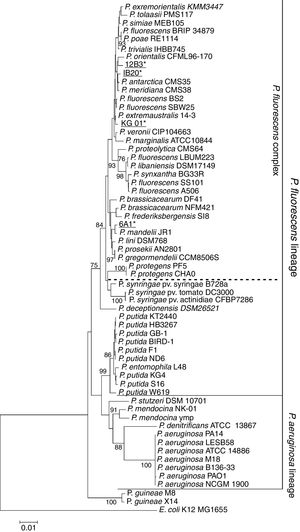
In order to determine the genus of the Antarctic bacterial isolates, we sequenced the 16S rRNA gene. The obtained sequences were deposited in GenBank (NCBI; Table S1). In a first step, the sequences were BLASTed against NCBI's 16S rRNA GenBank.21 For isolates IB20, 12B3 and KG01, sequence identity with any species contained in the database was below 97%, suggesting that all three isolates could represent new bacterial species.28 However, querying the sequence of strain 6A1 revealed a 99.9% sequence identity with the strain P. mandelii JR-1, meaning that the isolate might in fact belong to that species. On the other hand, the phylogenetic tree (Fig. 1) shows all of these strains as part of the P. fluorescens lineage and within that, the P. fluorescens complex, one of the most diverse and ubiquitous lineages of the genus.29 With regards to the other bacteria of the genus Pseudomonas that have been isolated in Antarctica, it can be seen that both strains of P. guineae form a lineage, which is separate from the ones that have been described for P. aeruginosa and P. fluorescens. However, these results are insufficient to draw conclusions.
Maximum-likelihood phylogenetic tree based on the partial 16S rRNA gene sequences of members of the Pseudomonas genus. The 16S sequence of E. coli K12 was used as an outgroup. The tree shows all bacteria of the genus Pseudomonas isolated in Antarctica to date (bold). Isolates presented in this study are shown in bold and underlined. It can be seen that the majority of the Antarctic bacteria are part of the P. fluorescens lineage, with the exception of the P. guineae isolates, which seem to be part of a separate lineage.
In order to expand on the results of the 16S rRNA gene analysis, we partially sequenced the five housekeeping genes aroE, glnS, gyrB, ileS and rpoD. Each one of these genes was then BLASTed against the GenBank nucleotide database (NCBI).21 The results are listed in Table 2. With the exception of the strain 6A1, where four of five genes show extremely high similarities (≤98%) demonstrating that this is the first strain of P. mandelii isolated in Antarctica; all three strains showed low similarities in at least four genes (≥96%). These results confirm that IB20, 12B3 and KG01 are serious candidates to be classify as a novel species of the Pseudomonas genus.
Percentage identity values obtained by BLASTing the five housekeeping genes of the four Antarctic isolates. The letters represent the different species and strains of Pseudomonas, to which the percentage identity refers.
| aroE | glnS | gyrB | ileS | rpoD | |
|---|---|---|---|---|---|
| IB20 | ≤90%g,h,i | ≤95%h,i,j | ≤94%i,k,l | ≤96%h,m,n | ≤99%o,p,q |
| 12B3 | ≤91%g,h,i | ≤96%h,i,j | ≤95%i,s,t | ≤96%h,m,n | ≤99%o,p,q |
| 6A1 | ≤94%a,b,c | ≤98%a,b,y | ≤98%a,b,d | ≤98%a,b,e | ≤99%a,b,f |
| KG01 | ≤88%m,u,v | ≤94%w,p,q | ≤94%i,x,w | ≤96%i,h,n | ≤99%p,q,w |
a=P. mandelii JR-1, b=P. fluorescens NCIMB11764, c=P. chlororapis subsp aurantiaca JD37, d=P. fluorescens PS1, e=P. mandelii DSM17967T, f=P. fluorescens FW300-N2E3, g=P. fluorescens SBW25, h=P. antarctica PAMC 27494, i=P. fluorescens LBUM 636, j=P. fluorescens UK4, k=P. reactans USB131, l=P. reactans USB20, m=P. trivialis IHBB745, n=P. fluorescens KENGFT3, o=P. fluorescens A506, p=P. fluorescens PICF7, q=P. fluorescens PCL1751, r=P. fluorescens NCIMB11764, s=P. yamanorum 8H1, t=P. putida MG2010, u=P. fluorescens L111, v=P. fluorescens L321, w=P. azotoformans S4, x=P. reactans USB94 y=P. koreensis D26.
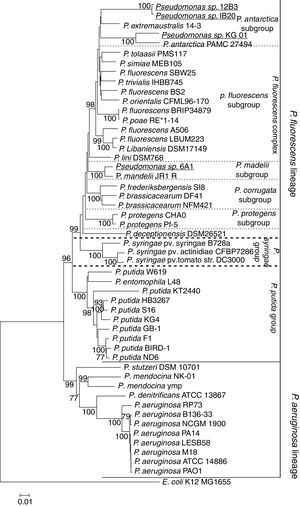
The sequences of the genes 16S, aroE, glns, gyrB, ileS and rpoD were concatenated to form a single sequence of 4150bp and compared against a concatenate of orthologous genes of the genus Pseudomonas. A concatenate of the same genes of E. coli strain K12 MG1655 was used as an outgroup. Sequences were aligned, and a phylogenetic tree was constructed using the maximum likelihood model with 1000 bootstraps replicas (Fig. 2). The two major lineages of the genus Pseudomonas, that of P. fluorescens and that of P. aeruginosa, can be clearly distinguished from one another. Likewise, within the P. fluorescens lineage, three subgroups can be clearly distinguished; these represent the P. fluorescens complex, the P. syringae group and the P. putida group. It can be seen that all Antarctic bacteria of the genus Pseudomonas described to date (in bold) are part of the P. fluorescens complex. Together with P. antarctica PAMC27494 and P. extremaustralis 14-3, the strains described here appear to form a new subgroup within this complex, which we have called the P. antarctica subgroup. The strain 6A1, on the other hand, forms a cluster with P. mandelii JR-1, confirming that it belongs to that particular species. Likewise, P. deceptionensis, though part of the P. fluorescens complex, belongs to another, as yet undescribed, subgroup.
Maximum-likelihood phylogenetic tree based on the concatenated sequences of the genes 16S-aroE-glnS-gyrB-ileS-rpoD, showing the evolutionary relationship among selected members of the Pseudomonas genus. The bacteria isolated in Antarctica are shown in bold, and isolates presented in this study are shown in bold and underlined. The tree clearly shows the lineages of P. aeruginosa and P. fluorescens. Likewise, the P. fluorescens complex and its subgroups can be clearly distinguished. All Antarctic Pseudomonas can be classified within the P. fluorescens complex. Among them, P. deceptionentis seems to form a separate subgroup, and the isolate Pseudomonas sp. 6A1 appears in a branch of the P. mandelii subgroup. Together with the species P. antarctica and P. extramaustralis, the isolates Pseudomonas sp. IB20, Pseudomonas sp. 12B3 and Pseudomonas sp. KG01 form the new subgroup P. antarctica.
The results of the MLSA analysis had already classified all strains as part of the P. fluorescens complex. To confirm this, a comprehensive phenotypic analysis of the Antarctic strains was carried out using BIOLOG GN2 and API 20 NE. The phenotypic analyses are summarized in Table 3. It is noteworthy that API 20 NE analyses classified all isolates within the species P. fluorescens. It is, however, known that the resolution of this technique is not particularly high, since it classifies most members of the P. fluorescens complex within this species.29 BIOLOG analyses yielded significant differences between the Antarctic isolates and the control strain P. fluorescens A506 (ATCC® 31948™). Based on these differences, three of the four new isolates can be considered good candidates for new species. The following specific findings are worth mentioning. The strain KG01 is the only one unable to assimilate d-mannose, l-proline, putrescine, pyruvic acid methyl ester, l-serine, and d,l-α-glycerol phosphate, while being the only strain able to assimilate adipic acid and p-hydroxyphenylacetic acid. IB20 is the only strain unable to assimilate d-arabitol and propionic acid; however, it is the only one able to assimilate l-phenylalanine (weakly), formic acid (weakly) and l-alaninamide. 12B3 is the only strain unable to assimilate malonic acid, urocanic acid and inosine. Finally, the strain 6A1 is the only one capable of assimilating N-acetyl-d-glucosamine, d-serine (weakly) and dextrin (weakly), while being unable to assimilate adonitol, d-galacturonic acid, d-glucuronic acid and l-ornithine. Additionally, this strain is unable to hydrolyze l-arginine and gelatin, thereby differing significantly from the other strains studied. Finally, all four Antarctic strains are able to assimilate d-trehalose, d-galactonic acid lactone, glycyl-l glutamic acid and d,l-carnitine, while the control, P. fluorescens A506 (ATCC 31948® ™), one of the most representative strains of the P. fluorescens complex, is not. Nevertheless, in the great majority of phenotypic tests, the results for the Antarctic strains are similar to those for the control, including the inability to assimilate nitrate, thereby corroborating that the Antarctic isolates are part of the P. fluorescens complex.
Phenotypic characterization of the Antarctic bacterial isolates. The phenotypic characterization was performed using BIOLOG GN2 and API 20 NE. +=positive test, −=negative test, W=weak reaction, a=API20NE, b=BIOLOG, *=P. fluorescens A506 (ATCC® 31948™).
| Test (Active ingredients) | Detail | KG01 | IB20 | 6A1 | 12B3 | P. fluorescens* |
|---|---|---|---|---|---|---|
| d-mannosea,b | Assimilation | − | + | + | + | + |
| l-Prolineb | Assimilation | − | + | + | + | + |
| l-Serineb | Assimilation | − | + | + | + | + |
| Putrescineb | Assimilation | − | + | + | + | W |
| Pyruvic Acid Methyl Esterb | Assimilation | − | + | + | + | W |
| d,l-α-Glycerol Phosphateb | Assimilation | − | + | W | + | − |
| d-Arabitolb | Assimilation | + | − | + | + | + |
| Propionic Acidb | Assimilation | + | − | W | + | + |
| Adonitolb | Assimilation | + | + | − | + | − |
| d-Galacturonic Acidb | Assimilation | + | + | − | + | - |
| d-Glucuronic Acidb | Assimilation | + | + | − | + | - |
| l-Ornithineb | Assimilation | W | + | − | + | - |
| Gelatine, bovine origena | Hydrolysis | + | + | − | + | - |
| Malonic Acidb | Assimilation | + | W | + | − | + |
| Urocanic Acidb | Assimilation | + | + | + | − | + |
| Inosineb | Assimilation | + | + | + | − | + |
| Adipic acida | Assimilation | + | − | − | − | − |
| p-Hydroxy Phenylacetic Acidb | Assimilation | + | − | − | − | + |
| l-Alaninamideb | Assimilation | − | + | − | − | − |
| l-Phenylalanineb | Assimilation | − | W | − | − | − |
| Formic Acidb | Assimilation | − | W | − | − | − |
| N-Acetyl-Dglucosamineb | Assimilation | − | − | + | − | + |
| d-Serineb | Assimilation | − | − | W | − | − |
| Dextrin | Assimilation | − | − | W | − | − |
| d-Trehaloseb | Assimilation | + | + | + | + | − |
| d-Galactonic Acid Lactoneb | Assimilation | + | + | + | + | − |
| Glycyl-Lglutamic Acidb | Assimilation | W | + | + | + | − |
| d,L-Carnitineb | Assimilation | + | W | + | + | − |
| Succinic Acid Mono-Methyl-Esterb | Assimilation | − | + | − | + | − |
| Succinamic Acidb | Assimilation | − | + | − | + | − |
| Glucuronamideb | Assimilation | − | + | − | + | − |
| Glycyl-Laspartic Acidb | Assimilation | − | + | − | W | − |
| i-Erythritolb | Assimilation | − | + | − | W | − |
| Xylitolb | Assimilation | − | W | − | + | − |
| Thymidineb | Assimilation | − | + | − | + | − |
| 2-Aminoethanolb | Assimilation | − | + | − | + | W |
| Tween 40b | Assimilation | − | + | + | + | + |
| L-Leucineb | Assimilation | − | + | W | + | W |
| d-Fructoseb | Assimilation | − | W | + | + | + |
| l-Threonineb | Assimilation | − | + | W | − | − |
| Hydroxy-l Prolineb | Assimilation | − | − | W | + | + |
| Uridineb | Assimilation | − | W | − | + | + |
| d-Sorbitolb | Assimilation | + | − | − | + | − |
In the present study, we describe four different Antarctic isolates, which we classified as part of the genus Pseudomonas using both molecular and phenotypic strategies. 16S rRNA gene analysis, MLSA and phenotypic tests using BIOLOG and API 20 NE all suggest that the Antarctic isolates Pseudomonas sp. KG01, Pseudomonas sp. IB20 and Pseudomonas sp. 12B3 are serious candidates to classify as new species of the genus. Likewise, we determined that the isolate Pseudomonas sp. 6A1 is the first strain of the species P. mandelii to be isolated in Antarctica.
There has been little research on the genus Pseudomonas on the Antarctic continent, and only eight species have been described in the past 20 years. Recent research has thoroughly revised the taxonomy and classification of the genus Pseudomonas,29 dividing it into two major lineages: that of P. aeruginosa, which includes almost exclusively bacteria isolated from clinical settings, and that of P. fluorescens, which contains most of the genus’ environmental bacteria, including the species P. putida and P. syringae, both of which have been widely studied. One of the most diverse groups within this lineage is called the “P. fluorescens complex”. It has been further subdivided into nine subgroups, P. protegens, P. chlororaphis, P. corrugata, P. koreensis, P. jessenii, P. mandelii, P. fragi, P. gessardii and P. fluorescens.29 All Antarctic strains studied to date have been classified within this complex. This is in line with our findings for the three candidates for new species, Pseudomonas sp. KG01, Pseudomonas sp. IB20 and Pseudomonas sp. 12B3. Likewise, the isolate Pseudomonas sp. 6A1, classified in this study as a strain of the species P. mandelii, also forms part of this complex. An original contribution of our work is that with these three potentially new species, we have identified a novel subgroup, which we have called P. antarctica, since it also incorporates the strains P. antarctica PAMC27494 and P. extremaustralis 14-3. These were previously considered to be part of the subgroup P. fluorescens.12 The hugely diverse P. fluorescens complex has a wide environmental distribution, and due to their versatile metabolism, most of its members play key roles at the ecological level.30,31 It is worth mentioning that some of the species of the P. fluorescens complex play an important role in biocontrol by producing secondary metabolites, including antibiotics and fungicides, which help protect plants from infections by fungi and pathogenic bacteria.32,33 In addition, it has been reported that certain subgroups of this complex can act in the bioremediation of heavy metals and pesticides.34 Furthermore, several members of the complex play a vital role in the carbon cycle.30 It has been suggested that the use of members of the subgroup P. fluorescens as plant growth-promoting rhizobacteria (PGPR) in plants of agronomical interest could reduce CO2 emissions, thereby mitigating global warming and the resulting climate change.35 On this background, the biotechnological potential of the three candidates for new species is obvious. Future studies are needed to analyze their possible use as plant growth-promoting rhizobacteria (PGPR), as well as their use in biocontrol and bioremediation.
It is worth noting that the four isolates were collected from different environments, including seawater, marine sediment and freshwater, suggesting that the P. fluorescens complex might have a notable capacity of colonization of diverse environments in the Antarctic continent. This does not seem to be uncommon in extreme environments, and a study from the Himalayas, another environment of extreme cold, also reported the P. fluorescens complex to be highly represented within the genus Pseudomonas.36 Of 336 specimens of the genus Pseudomonas, 308 isolates belonged to this complex. Of note, the temperatures of the sampling sites ranged from 2 to 11°C in the Himalayas, thus confirming that members of this complex are tolerant of the cold. Future studies examining a large number of Antarctic isolates are needed to determine the abundance of this complex on the Antarctic continent and to elucidate the ecological role played by these bacteria.
Due to the high volume of whole genome sequencing data and metagenomic analyses, the description of new bacterial species is turning increasingly more complex. MLSA is a robust technique to determine whether a particular isolate belongs to a previously described species, or whether it represents a new species. Several studies have shown that the study of concatenated sequences of carefully selected housekeeping genes is a powerful and reliable tool to identify new species within a genus.37,38 Here, we suggest that, in addition to 16S-rRNA, the genes aroE, gyrB, ileS, glnS and rpoD are ideal for use in MLSA in the genus Pseudomonas, since they combine all necessary features: they are “housekeeping” genes, they are protein-coding, and they are not transmitted horizontally.37,39 Although previous studies have suggested that the gene ileS is not a good candidate to be used in MLSA,40 in this study, it proved to be highly efficient. On the other hand, the gene rpoB is commonly used in MLSA;29,41–44 however, despite trying several sets of primers, we failed to obtain a unique product suitable for sequencing from our Antarctic bacterial isolates. The gene was therefore excluded from the analyses. One of the strengths of our work is that we performed MLSA on a concatenate of six genes (16S, aroE, gyrS, glnS, ileS and rpoD), as opposed to the three or four genes used in most studies characterizing members of the genus Pseudomonas. This resulted in a significantly more robust analysis. Without doubt, one of the main problems of this study lay in the shortage of available Antarctic Pseudomonas sequences. Sequences for all genes (aroE, glnS, gyrB, ileS and rpoD) were only available for the strains P. antarctica PAMC27494, P. extremaustralis 14-3 and P. deceptionensis DSM26521. The phylogenetic tree obtained through MLSA could therefore be further complemented when the sequences of other species become available in the database. For the 16S rRNA gene, however, sequences from all Antarctic species were available.
Molecular identification by MLSA has proven to be an effective tool to determine whether a bacterial isolate belongs to a particular species. However, in addition to phylogenetic, phenotypic or serological analyses, the most recommended method to discriminate between bacterial species, and to characterize new bacterial species, is DNA-DNA hybridization.45 This method does, however, have limitations: it takes long to develop and relies on specialized expertise, it does not define the phylogenetic distance between species, and it is not cumulative.42 A fast and robust method of classification for the genotypic characterization based on the sequences of protein-coding genes, MLSA has allowed researchers to classify a diverse and previously undescribed group of prokaryotes obtained from metagenomics analyses, many of which cannot be cultured.28,42 It has therefore even been argued that DNA-DNA hybridization analysis may no longer be necessary for the description of a new species.42,43,46
In conclusion, our results strongly suggest that the Antarctic isolates Pseudomonas sp. KG01, Pseudomonas sp. IB20 and Pseudomonas sp. 12B3 are candidates for new species of the genus Pseudomonas, that all of them belong to the P. fluorescens complex, and that, together with the Antarctic strains P. antarctica PAMC27494 and P. extremaustralis 14-3, they form part of the new subgroup “P. antarctica”. Future studies including metagenomics studies, are needed to determine the abundance of members of the genus Pseudomonas in Antarctica and to elucidate their possible ecological role on the cold continent.
FundingThis work was supported by the Dirección de Investigación of the Pontificia Universidad Católica de Valparaíso [Project DI 037.431/2015], by an INACH PhD scholarship [DT_02-13], and by the Programa de Investigación Asociativa [PIUAs-PUCV 037.293-2015].
Conflicts of interestThe authors declare that they have no conflict of interest.
We would like to thank Dr. Marcelo González of the Chilean Antarctic Institute (INACH) for providing the Antarctic bacterial isolates.