Erythrina velutina (“mulungu”) is a legume tree from Caatinga that associates with rhizobia but the diversity and symbiotic ability of “mulungu” rhizobia are poorly understood. The aim of this study was to characterize “mulungu” rhizobia from Caatinga. Bacteria were obteined from Serra Talhada and Caruaru in Caatinga under natural regeneration. The bacteria were evaluated to the amplification of nifH and nodC and to metabolic characteristics. Ten selected bacteria identified by 16S rRNA sequences. They were tested in vitro to NaCl and temperature tolerance, auxin production and calcium phosphate solubilization. The symbiotic ability were assessed in an greenhouse experiment. A total of 32 bacteria were obtained and 17 amplified both symbiotic genes. The bacteria showed a high variable metabolic profile. Bradyrhizobium (6), Rhizobium (3) and Paraburkholderia (1) were identified, differing from their geographic origin. The isolates grew up to 45°C to 0.51molL−1 of NaCl. Bacteria which produced more auxin in the medium with l-tryptophan and two Rhizobium and one Bradyrhizobium were phosphate solubilizers. All bacteria nodulated and ESA 90 (Rhizobium sp.) plus ESA 96 (Paraburkholderia sp.) were more efficient symbiotically. Diverse and efficient rhizobia inhabit the soils of Caatinga dry forests, with the bacterial differentiation by the sampling sites.
Caatinga (etymology: white forest) is the main phytophysiognomy occurring in Brazilian Semi-Arid region, occupying more than 70% of Brazilian Northeastern. These lands encompasses several dry forests that shows, as the main characteristics, low rainfall (below 800mmyr−1) concentrated in the firsts 3–4 months of the year and high temperature averages.1 The plant biodiversity in Caatinga is very high with the predominance of Fabaceae (Leguminosae) family, with 82 genera and 617 already cataloged species.2 The historical use of lands in Caatinga for shifting agriculture practices leaded to several lands in fallow, where the natural regeneration occur. Nowadays Caatinga dry forests are a great mosaic of fallow lands with different stages of natural regeneration.3,4 In this process, plants belonging to Fabaceae are the pioneers colonizers and can present an important ecological role.3–6
Several leguminous pioneers colonizing the Caatinga natural regeneration lands are able to associate with indigenous rhizobia from the soils of the region. Rhizobia are root and/or stem nodulating, nitrogen-fixing, classified within α and β-proteobacteria subclasses. They hold the nitrogenase enzymatic complex, able to reduce the atmospheric N2 to ammonium. The biological nitrogen fixation (BNF) is the main source of nitrogen incorporation in the food webs, mainly by the association between legumes and rhizobia.7 BNF is exploited in agriculture and forestry by the production and application of rhizobial inoculants that reduces the costs and environmental impacts of plant production, in addition to yield increase.8 The continuous selection of new and more efficient rhizobial strains is important to obtain bacteria with higher agronomic performance and agricultural applications.8,9 In natural regeneration lands, the selection of native and adapted rhizobia can help to improve the regeneration process due to their use in the production and application of more adapted and healthy seedlings.
Besides the nitrogen fixation, rhizobia are able to directly induce the plant growth by other mechanisms, such as the phytohormone production (auxin, for example) and release of soil insoluble nutrients (e.g. phosphorus and iron) among several others.10,11 These mechanisms can easily be in vitro evaluated by simple and low costs techniques, showing correlation with the bacterial performance in plant inoculation experiments.12,13 At the same way, the evaluation of rhizobial tolerance to environmental stresses, such as the salinity and temperature tolerance, also can be easily performed in vitro14 and can be correlated to the results of plant inoculation experiments.13 The evaluation of in vitro environmental stresses are important to the selection of bacteria that are (likely) more tolerant and also to better understand the bacteria biology.15 These features are particularly interesting to be evaluated in studies of rhizobia from tropical dry lands with low rainfall and high temperature averages.
Recent studies showed that Erythrina velutina Willd., commonly known as “mulungu”, is able to establish symbiotic association with rhizobia in soils from the Brazilian Semi-Arid.16,17E. velutina, classified within the Papilionoidae clade and Phaseoleae tribe, is a naturally inhabiting tree from the Northeast region of Brazil. This species is used as wood, timber, ornamental applications and sources of bioactive compounds with pharmacological applications.18,19 In addition, due to the fast growth and adaptive characteristic to the Semi-Arid environment, “mulungu” seedlings should be applied in land regeneration projects.20
Despite the potential use of this native species for several applications, there are not official recommendations of rhizobial strains for E. velutina in Brazil. The selection of native nitrogen-fixing isolates can help to the inclusion of the “mulungu” bacteria in the list of officially recommended strains to inoculant production by the Brazilian Ministry of Agriculture, Livestock and Food Supply (MAPA). The application of selected efficient rhizobia is useful to the development of improved “mulungu” seedlings, more healthy and with better nutrition, increasing the probability of the successful establishment in the fields, especially in non-favorable environments, such as the Caatinga degraded lands.21 Thus, we hypothesized that the Caatinga soils under natural regeneration in the Semi-Arid region of Pernambuco State harbor a diversity of efficient “mulungu” rhizobia. The aim of the present study was to characterize the “mulungu” rhizobia from the semi-arid region of Pernambuco State regarding their phenotypical, molecular and symbiotic characteristics.
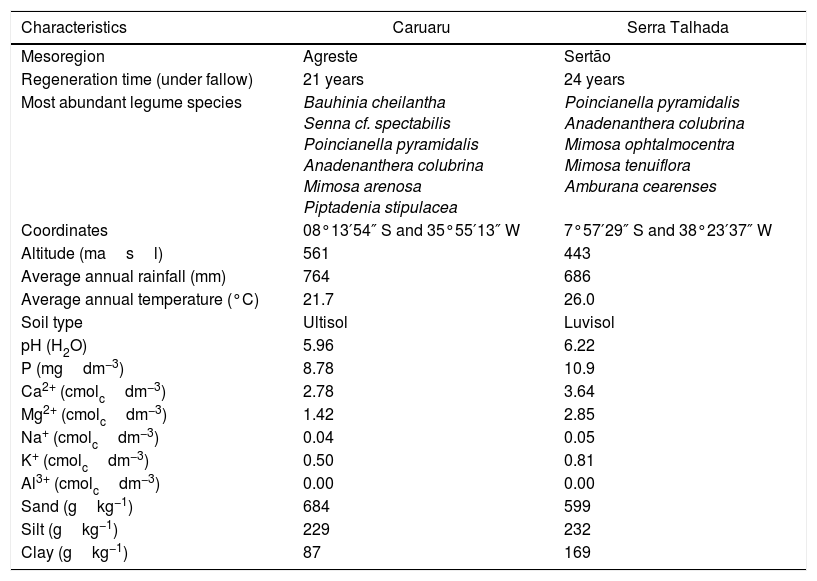
Materials and methodsOrigin of the bacterial isolatesThe bacteria were isolated from soils collected from the 0 to 20cm superficial layer in areas of Caatinga vegetation under natural regeneration in Caruaru, in the Agreste zone of Pernambuco state, and Serra Talhada, in the Sertão zone of the same state. The plant cover an both zones are different as already pointed by Silva and Rodal.22 The location and edaphoclimatic conditions of both areas are described in Table 1.
Characteristics of Caatinga under natural regeneration and their soils in Caruaru and Serra Talhada (Pernambuco State, Brazil).
| Characteristics | Caruaru | Serra Talhada |
|---|---|---|
| Mesoregion | Agreste | Sertão |
| Regeneration time (under fallow) | 21 years | 24 years |
| Most abundant legume species | Bauhinia cheilantha Senna cf. spectabilis Poincianella pyramidalis Anadenanthera colubrina Mimosa arenosa Piptadenia stipulacea | Poincianella pyramidalis Anadenanthera colubrina Mimosa ophtalmocentra Mimosa tenuiflora Amburana cearenses |
| Coordinates | 08°13′54″ S and 35°55′13″ W | 7°57′29″ S and 38°23′37″ W |
| Altitude (masl) | 561 | 443 |
| Average annual rainfall (mm) | 764 | 686 |
| Average annual temperature (°C) | 21.7 | 26.0 |
| Soil type | Ultisol | Luvisol |
| pH (H2O) | 5.96 | 6.22 |
| P (mgdm−3) | 8.78 | 10.9 |
| Ca2+ (cmolcdm−3) | 2.78 | 3.64 |
| Mg2+ (cmolcdm−3) | 1.42 | 2.85 |
| Na+ (cmolcdm−3) | 0.04 | 0.05 |
| K+ (cmolcdm−3) | 0.50 | 0.81 |
| Al3+ (cmolcdm−3) | 0.00 | 0.00 |
| Sand (gkg−1) | 684 | 599 |
| Silt (gkg−1) | 229 | 232 |
| Clay (gkg−1) | 87 | 169 |
For bacterial isolation, a trap-host pot experiment was implemented using “mulungu” (E. velutina) as trap plant. Polystyrene pots (500mL) were filled with the soil samples and four seeds were sowed per pot. Ten days after the emergence (DAE) a single plant was left per pot. The pots were daily irrigated with 100mL of distilled water and the experiment was harvested at 85 DAE. The roots and shoots were separated and the nodules detached. For isolation, nodules were superficially disinfected with 96°GL ethanol for 30s; sodium hypochlorite (2.5%, v/v) for five minutes followed by 10 washes in sterile distilled water. The nodules were crushed in yeast extract-mannitol-agar (YMA) medium with congo red23 and incubated in a growth chamber at 28°C. The rise of typical rhizobial colonies were daily evaluated during ten days. The colonies were purified in YMA medium with bromothymol blue and stored in YM medium+glycerol (2.5% v/v) at −80°C and deposited in the Culture Collection of Agricultural Interests Micro-organisms at Embrapa Semiárido (CMISA).
Amplification of symbiotic genesThe bacterial isolates were grown in liquid YM medium for three days for the fast-growing isolates and six days for the slow-growing. An aliquot of 1mL of each broth was used for the DNA extraction using the Wizard® Genomic DNA Purification System (Promega, USA) according to the manufacturer's instructions. nifH and nodC genes were co-amplified in a duplex-PCR reaction as described by Fernandes Júnior et al.24 For the nifH, the primers PolF (TGCGAYCCSAARGCBGACTC) and PolR (ATSGCCATCATYTCRCCGGA)25 were used. For nodC, the primers NodCF (AYGTHGTYGAYGACGGTTC) and NodCR(I) (CGYGACAGCCANTCKCTATTG)26 were applied. For a single isolate that showed positive amplification of the nifH amplicon, a complementary uniplex-PCR was performed with the primers nodCForB (CTCAATGTACACARNGCRTA) and nodCRevB (GAYATGGARTAYTGGYT)57 targeting the nodC amplification of β-rhizobia.
The reactions were performed in a Veriti 96-well thermocycler (Applied Biosystems, USA) and the PCR products were submitted to horizontal electrophoresis in agarose gel (0.8%, w/v). The gel was stained with Gel Red (Biotium) and visualized in a UV chamber. The bacterial isolates positive for both amplicons were selected for the next steps.
Metabolic characterization using the API 20 NE® kitEnzymatic activities and the carbon sources utilization profiles were performed using the API 20 NE® strips (BioMérieux, France) according to the manufacturer's instructions. The incubation time was three days for the fast-growing isolates, and six days for the slow-growing, at 28°C.
The results were transformed in a binary matrix and the bacteria clustered according to the enzymatic activity characteristics and utilization of different carbon sources in a similarity dendrogram obtained applying the Jaccard similarity coefficient and the UPGMA clustering algorithm with the aid of the BioNumerics v. 7.5 software package (Applied Maths, Belgium). The clustering analysis was used to select the bacterial isolates for further evaluations.
16S rRNA gene sequence analysesThe 16S rRNA gene was amplified using the universal primers 27F (AGAGTTTGATCMTGGCTCAG) and 1492R (TACGGYTACCTTGTTACGACTT).27 To evaluate the success of PCR, the products were submitted to a gel electrophoresis as described above. The PCR products were purified with the Wizard® SV Gel and PCR Clean-Up System (Promega, USA) commercial kit, following manufacturer's instructions. The products were sequenced with both forward and reverse primers in a 3730 xl genetic analyzer (Applied Biosystems, USA) at Macrogen (Seoul, South Korea).
The quality of the sequences was verified using the Sequence Scanner Software v. 2.0 (Applied Biosystems, USA). Good quality sequences were used to construct the contigs for the bacterial identification through comparison with the sequences available at the EzBioCloud database (http://www.ezbiocloud.net/).28 Sequences from closest type strains were downloaded and used to perform the phylogenetic tree. The alignment of the sequences was carried out by MUSCLE and the Neighbor-Joining tree made with the Jakes-Cantor model using the bootstrap phylogeny test with 1000 replications. The sequence alignments and phylogenetic tree construction were carried out in MEGA 6.0 software.29 The sequences were deposited in the GenBank, database of the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/Genbank), under the accession numbers MF288754 to MF288763.
Salt and temperature toleranceStrains were analyzed for salt and high temperature tolerance, following the methodology described by Fernandes Júnior et al.14 with some modifications, briefly described below. To the evaluation of salt tolerance, the strains were inoculated in Petri dishes with YMA medium supplemented with zero (control), 0.085, 0.17, 0.34 and 0.51molL−1 of NaCl and incubated in a growth chamber at 28°C. To evaluate the growth ability of the strains submitted to different incubation temperatures the isolates were inoculated in Petri dishes containing original YMA medium and incubated at 28 (control), 35, 40 and 45°C, in different growth chambers. In both experiments, conducted within a completely randomized design with three replications, the strains were incubated for three and six days for the fast-growing and slow-growing, respectively.
Auxin production and solubilization of calcium phosphateThe in vitro production of auxin was evaluated according to the colorimetric method described by Sarwar and Kremer30 with modifications, briefly described below. The strains were grown in YMA medium and were checked for purity. Pure colonies were inoculated in YM medium (3mL), and after three days for the fast-growing isolates and six days for the slow growing, the bacterial broth were adjusted to OD540=0.5 with distilled autoclaved water (DAW). An aliquot of 1mL of each adjusted broth were centrifuged (6000×g for 5min), the supernatant discarded and pellet re-suspended with 1mL of distilled autoclaved water.
To evaluate the metabolic pathways enrolled in the in vitro auxin production, aliquots of 150μL of the re-suspended bacteria were inoculated in standard liquid YM culture medium, without bromothymol blue, supplemented or not with 168mgL−1 of l-tryptophan (l-try). The media were incubated at 28°C (±1°C) in an orbital shaker with constant stirring at 120rpm, for four and seven days to the fast and slow-growing bacteria, respectively. After the incubation period, the cultures were adjusted to an OD540=0.5 as described above. Aliquots of standardized bacterial solutions (1mL) were centrifuged at 6.000×g for 5min. Aliquots of 200μL of supernatant were placed in 96-well ELISA microplates and mixed with 100μL of Salkowski solution (1mL of FeCl3 0.5molL−1+49mL of HClO4 6molL−1). The ELISA plates were incubated in the dark for 30min. The intensity of red color was determined in a MultiSkan GO spectrophotometer (Thermo Scientific, Germany) at 530nm. The concentration of auxin was estimated using a standard curve previously prepared with a range of 0 to 500μgL−1 of synthetic indole-3-acetic acid (Sigma–Aldrich, USA).
The isolates were evaluated for their capacity to in vitro solubilize calcium phosphate in the solid GL medium supplied with insoluble CaHPO4.31 The bacteria were grown in liquid YM medium, OD adjusted, centrifuged, and re-suspended as described above. Three aliquots of 10μL were dropped in equidistant points in the center of the Petri dishes. The bacteria were incubated at 28°C for seven days for the fast-growing and fifteen days for the slow growing. After the incubation period, the diameter of the colonies and the translucent zone surrounding the colonies were measured in millimeters (mm) with a ruler. This data were used to calculate the Solubilization Index (SI)=diameter of translucent zone/diameter of colony.32
The experiments of in vitro auxin production and calcium phosphate solubilization were performed in a completely randomized design with three replications.
Symbiotic efficiency in “mulungu” plantsThe symbiotic efficiency of the isolates were evaluated under gnotobiotic conditions in a greenhouse. The experiment was carried out at the Embrapa Semiárido facilities in Petrolina, Pernambuco state. For this assay, the dormancy of “mulungu” seeds was broken by mechanical scarification of the tegument. The seeds were surface disinfected with ethanol 96° GL for 30s, sodium hypochlorite 2.5% (v/v) for five minutes, followed by eight washes with DAW.23 The substrate was washed and sterilized sand (autoclaved twice, at 120°C and 1.5atm for 1h, with 72h between the sterilizations). The experiment was set up in polystyrene pots (500mL), which were disinfected by washing with sodium hypochlorite 2.5% (v/v), followed by three washes with DAW. Pots were carefully filled with the sterile sand and four seeds per pot were sown soon after filling.
For inoculation, bacteria were grown in YM medium up to the end of the exponential growth phase (around 109cellsmL−1), for three days for the fast-growing strains and six days for the slow-growing, in an orbital shaker at 28°C, as described above. Right after sowing, the inoculation was performed by the drop of 2mL of the bacterial broth on each seed. At twenty DAE a thinning was performed and a single plant was left per pot. The pots were daily supplied with 100mL of DAW, and after the cotyledon drop (around 25–28 DAE), 50mL of nitrogen free nutrient solution, described by Norris and T’Mannetje,33 was applied once a week.16
The experimental treatments consisted of the inoculation of ten bacteria (single inoculation of each isolate), a positive control inoculated with Bradyrhizobium elkanii BR 5609 (SEMIA 6100), strain officially recommended by MAPA in Brazil for use as inoculant for Erytrina verna and Falcataria mollucanna, and two uninoculated controls: one supplied with NH4NO3 (70mg N plant−1 week−1) applied after 35 DAE; and one without nitrogen supplementation.
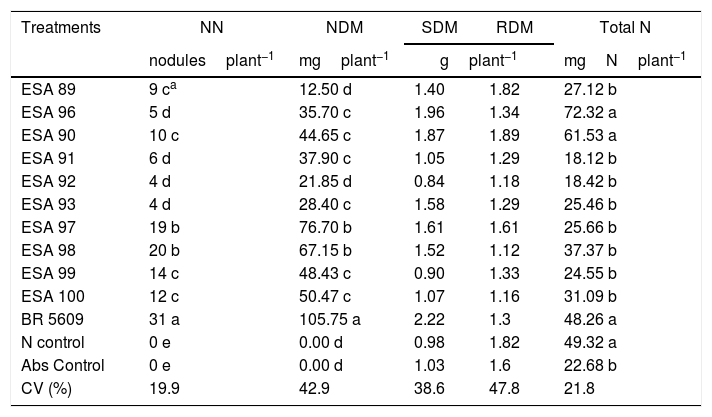
The plants were harvested at 92 DAE for the determination of the nodule number (NN), nodule dry matter (NDM), shoot dry matter (SDM), root dry matter (RDM) and nitrogen accumulation in the shoot (total N). For the NN evaluation, the nodules were detached from the roots and counted. For the NDM, SDM and RDM, respectively, the nodules, shoots and roots were separately in paper bags and dried in a forced-air oven at 65°C for seven days and weighted. The shoots were grounded for determination of shoot nitrogen concentration by the dry combustion method in a TruSpec CN elemental analyzer (Leco, USA). These values were used for calculation of total accumulation of nitrogen in the shoot (Total N) through the multiplication of the nitrogen concentration by the SDM.
Statistical analysesFor the solubilization of calcium phosphate, IAA production, and symbiotic efficiency in greenhouse experiments, the normal distribution of the errors were evaluated by the Shapiro–Wilk test, thus the data for the greenhouse experiment were transformed by (x+1)0.5 to reach the normal distribution.
The experimental data were submitted to variance analysis (ANOVA) and the averages were compared by the Scott–Knott's mean range test (p<0.05). The data were analyzed using the statistical package Sisvar v 5.0.34
ResultsIsolation and symbiotic gene amplificationThe isolation process retrieved 32 bacteria, 17 from Caruaru soils and 15 from Serra Talhada. The duplex-PCR reaction to nifH and nodC resulted in the positive reaction for both genes to seven (47%) bacterial isolates from Serra Talhada and 10 (59%) bacteria from Caruaru, including ESA 96, a single fast-growing isolate that did not amplify the nodC at the duplex-PCR reaction but amplified the nodC when a primer pair to β-rhizobia was applied.
Among these bacteria, ten were slow (seven from Caruaru and three from Serra Talhada) and seven were fast-growing (four from Caruaru and three from Serra Talhada), respectively. All 15 isolates that did not amplify the symbiotic genes were fast-growing bacteria.
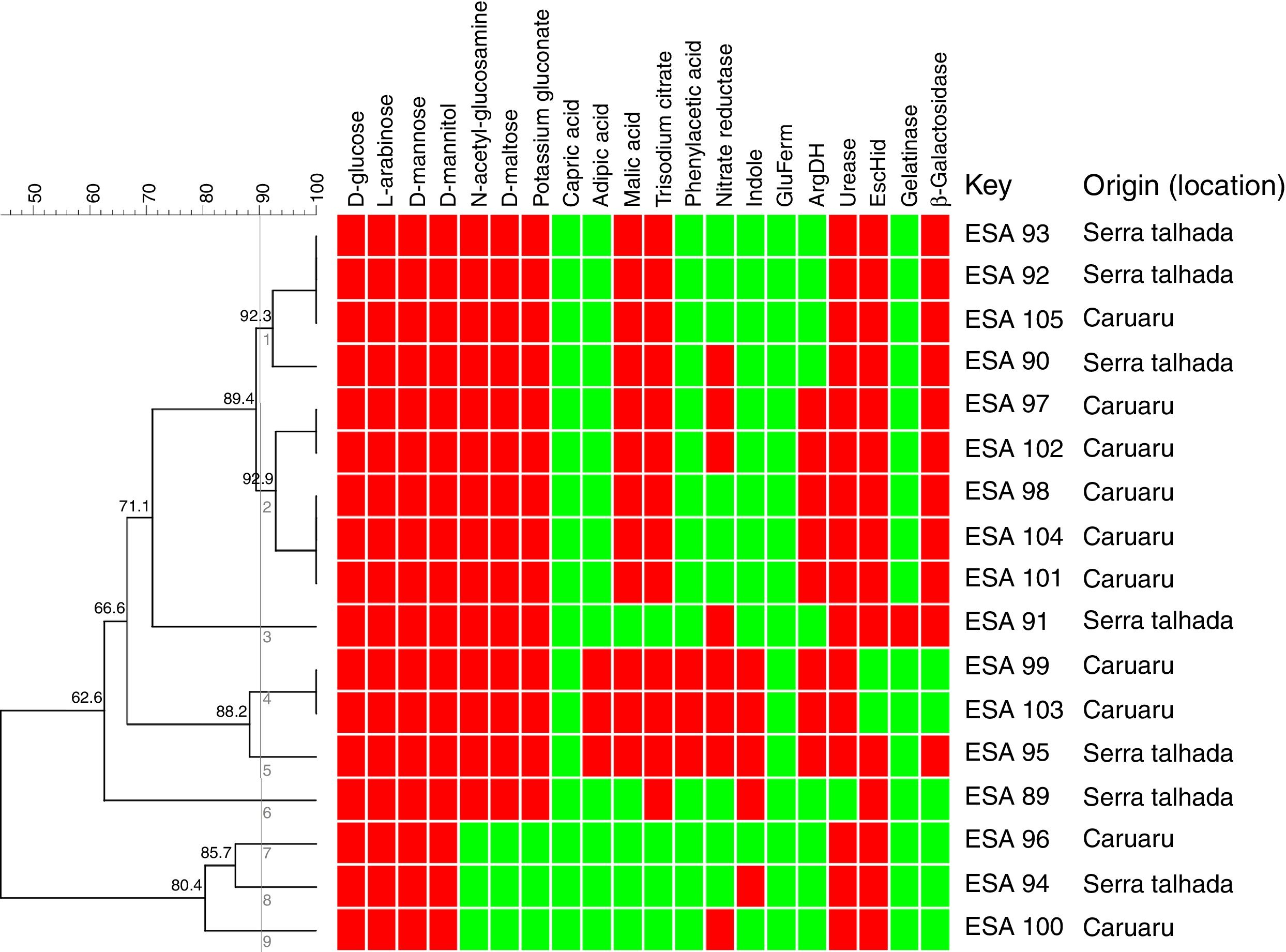
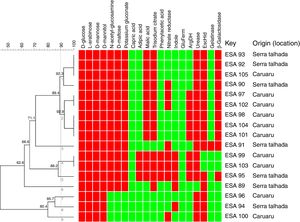
Metabolic profile of the bacterial isolatesRegarding metabolism of the 12 carbon sources available at the API 20 NE strips, a large variability at the metabolic profile were observed in the 17 bacteria evaluated. All bacterial isolates grew in d-glucose, l-arabinose, d-mannose and d-mannitol, as sole carbon sources. None of the bacteria grew using capric acid. Around 83% grew with N-acetyl-glucosamine, d-maltose and potassium gluconate. Around 76 and 70% used respectively, trisodium citrate and malic acid as sole carbon sources. The bacteria ESA 95, ESA 103 and ESA 99 showed the best ability to metabolize the carbon sources since they grew in all sources, excepting the capric acid. These bacteria were also the only isolates that grew using adipic acid and phenylacetic acid (Fig. 1).
Cluster analysis of the metabolic profile of 17 bacterial isolates of Erythrina velutina based on the results of 12 carbon sources assimilation and 8 enzymes activities in the API 20 NE strips. First 12 columns: carbon sources metabolism. Last 8 columns: Enzymatic activity. Indole, indole formation; GluFerm, glucose fermentation; ArgDH, arginine dehydrogenase. Red squares: positive activity; Green squares: negative activity. Numbers in the nodes are the cophenetic correlation. Key, Bacterial isolate.
For the enzymatic activity, the isolates showed different profiles. All bacterial isolates did not presented activity of glucose fermentation. Excepting the isolate ESA 89, all bacterial isolates were positive to urease and, excepting the ESA 99 and ESA 103, all isolates were positive to esculin hydrolysis. An amount of 65% of the bacterial isolates was positive to β-Galactosidase. Variable profiles were achieved from the evaluations of nitrate reductase, indol formation, arginine DiHydrolase, and gelatinase.
All metabolic results were tabulated in a binary matrix and used to the cluster analysis of all bacteria. The similarity dendrogram (Fig. 1) presents the formation of nine clusters at the threshold of 90% (Jaccard coefficient). Based on the cluster analysis, ten bacterial isolates were selected for further steps.
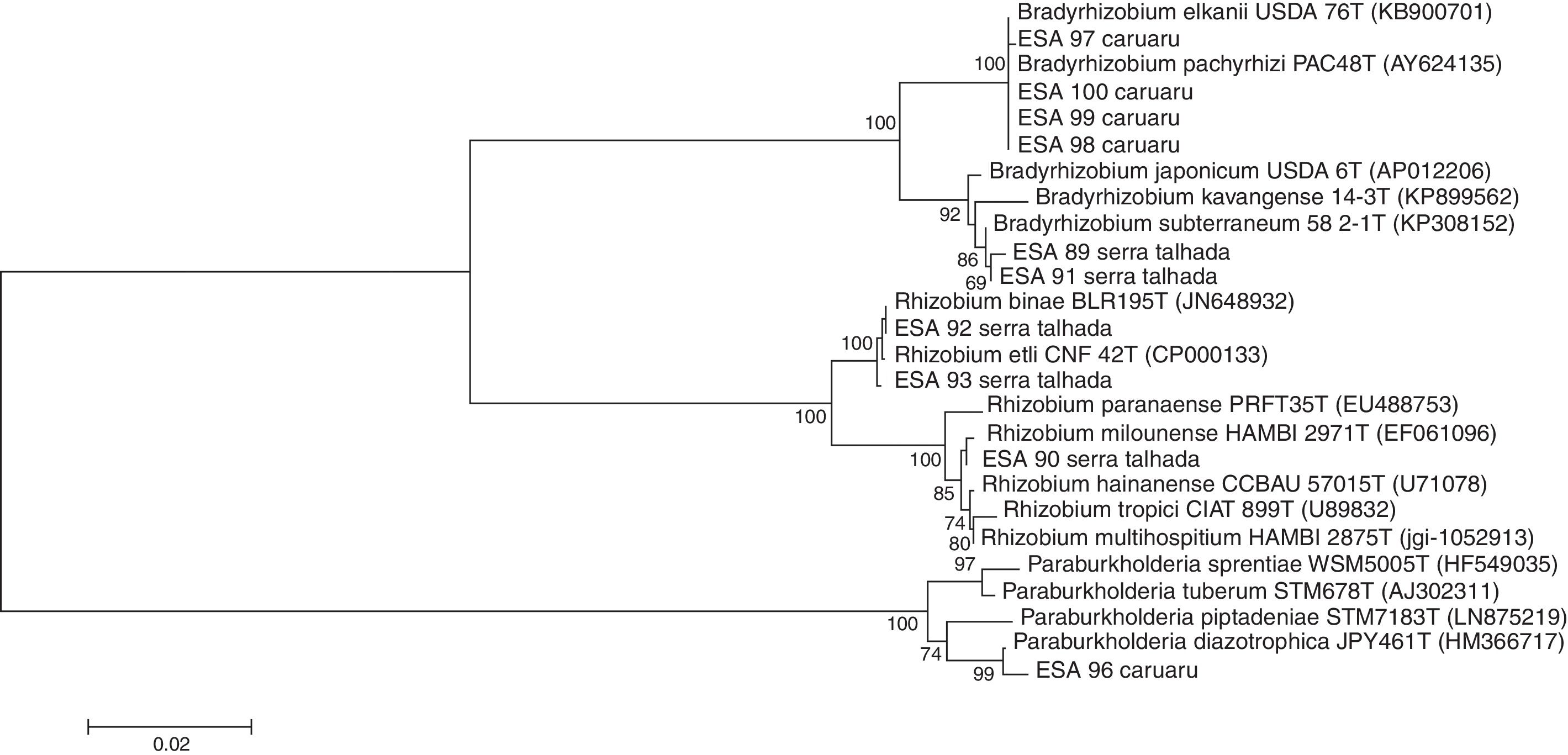
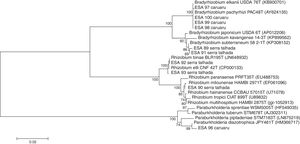
16S rRNA gene sequence analysesThe sequences of 16S rRNA gene were compared with those available at the EzBioCloud database. The comparison indicated that among the five bacterial isolates from Caruaru, four of them were classified as α-rhizobia and clustered within the Bradyrhizobium in the B. elkanii clade II. The isolate ESA 96 was classified as a β-rhizobia related to Paraburkholderia diazotrophica. The five bacteria from Serra Talhada were classified as α-rhizobia. The isolates ESA 92 and ESA 93 were classified within Rhizobum in the R. etli clade. The bacteria ESA 90 was classified in the same genus but in the R. tropici clade. The isolates ESA 89 and ESA 91 were classified within the Bradyrhizobium japonicum clade, closely related to Bradyrhizobium subterraneum (Fig. 2).
Neighbor-joining phylogenetic tree using a Jukes-Cantor model based on the partial 16S rRNA gene sequence (1165 nt) of ten rhizobial isolates from root nodules of Erythrina velutina and 16 type strains. Numbers in the nodes of branches correspond to the bootstrap value from 1000 replications. Bootstrap values lower than 60% are not shown.
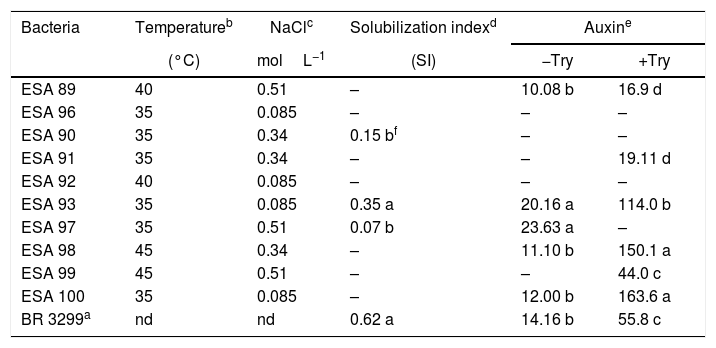
Two isolates (ESA 89 and ESA 92) grown under the incubation temperature of 40°C. The bacterial isolates ESA 98 and ESA 99 grew when incubated at 45°C. The other bacterial isolates grew only under the incubation temperature of 35°C (Table 2). For the tolerance to different NaCl concentrations, the bacteria grew in the YMA medium supplemented with 0.085–0.51molL−1 of NaCl. The isolates ESA 90, ESA 91 and ESA 98 grew positively in the medium supplemented with NaCl 0.34molL−1 while the bacteria ESA 89, ESA 97 and ESA 99, grew in the medium with NaCl 0.51molL−1. The isolates ESA 100, ESA 93, ESA 92 and ESA 96 were the less tolerant to salinity and grew only with the lower NaCl concentration in the medium (0.085molL−1).
In vitro temperature and NaCl tolerance, calcium phosphate solubilization and auxin production of ten rhizobial isolates from Erythrina velutina root nodules, obtained from “Caatinga” dry forest soils, in Pernambuco state, Brazil.
| Bacteria | Temperatureb | NaClc | Solubilization indexd | Auxine | |
|---|---|---|---|---|---|
| (°C) | molL−1 | (SI) | −Try | +Try | |
| ESA 89 | 40 | 0.51 | – | 10.08 b | 16.9 d |
| ESA 96 | 35 | 0.085 | – | – | – |
| ESA 90 | 35 | 0.34 | 0.15 bf | – | – |
| ESA 91 | 35 | 0.34 | – | – | 19.11 d |
| ESA 92 | 40 | 0.085 | – | – | – |
| ESA 93 | 35 | 0.085 | 0.35 a | 20.16 a | 114.0 b |
| ESA 97 | 35 | 0.51 | 0.07 b | 23.63 a | – |
| ESA 98 | 45 | 0.34 | – | 11.10 b | 150.1 a |
| ESA 99 | 45 | 0.51 | – | – | 44.0 c |
| ESA 100 | 35 | 0.085 | – | 12.00 b | 163.6 a |
| BR 3299a | nd | nd | 0.62 a | 14.16 b | 55.8 c |
The isolates ESA 90, ESA 93 and ESA 97, were able solubilize calcium phosphate, highlighting the isolate ESA 93, statistically superior (p<0.05) than the other two bacteria. Six and five isolates produced auxin in the presence and absence of l-try supplementation, respectively. In the medium with l-try, ESA 100 and ESA 98 were in the highest cluster in the mean range test comparison. This group was followed by the isolate ESA 93 that was higher than other group with the isolate ESA 99 and the reference strain BR 3299T. Considering the auxin production in YM medium without l-try, the isolates ESA 93 and ESA 97 showed the best performance, comparing to the other isolates and the reference strain.
Symbiotic efficiency in “mulungu” plantsAt the symbiotic efficiency experiment in gnotobiotic conditions, all replications of the non-inoculated controls (both the absolute and the nitrogen supplied controls) did not nodulate, being inferred that contaminations did not occur in the experiment. The positive control inoculated with the reference strain of B. elkanii, and the other 10 treatments inoculated with the new bacterial isolates induced nodule formation in “mulungu” roots (Table 3).
Averages for the nodule number (NN), nodule dry matter (NDM), shoot dry matter (SDM), root dry matter (RDM), and accumulation of nitrogen in the shoot (total N) of Erythrina velutina inoculated with new rhizobial isolates in a gnotobiotic conditions experiment.
| Treatments | NN | NDM | SDM | RDM | Total N |
|---|---|---|---|---|---|
| nodulesplant−1 | mgplant−1 | gplant−1 | mgNplant−1 | ||
| ESA 89 | 9 ca | 12.50 d | 1.40 | 1.82 | 27.12 b |
| ESA 96 | 5 d | 35.70 c | 1.96 | 1.34 | 72.32 a |
| ESA 90 | 10 c | 44.65 c | 1.87 | 1.89 | 61.53 a |
| ESA 91 | 6 d | 37.90 c | 1.05 | 1.29 | 18.12 b |
| ESA 92 | 4 d | 21.85 d | 0.84 | 1.18 | 18.42 b |
| ESA 93 | 4 d | 28.40 c | 1.58 | 1.29 | 25.46 b |
| ESA 97 | 19 b | 76.70 b | 1.61 | 1.61 | 25.66 b |
| ESA 98 | 20 b | 67.15 b | 1.52 | 1.12 | 37.37 b |
| ESA 99 | 14 c | 48.43 c | 0.90 | 1.33 | 24.55 b |
| ESA 100 | 12 c | 50.47 c | 1.07 | 1.16 | 31.09 b |
| BR 5609 | 31 a | 105.75 a | 2.22 | 1.3 | 48.26 a |
| N control | 0 e | 0.00 d | 0.98 | 1.82 | 49.32 a |
| Abs Control | 0 e | 0.00 d | 1.03 | 1.6 | 22.68 b |
| CV (%) | 19.9 | 42.9 | 38.6 | 47.8 | 21.8 |
No differences were found for the variables SDM and RDM, in all treatments. For the nodulation variables, NN and NDM indicated that the reference strain BR 5609 was higher than the other treatments. Considering the new bacteria, the isolates ESA 97 and ESA 98 stood out, showing higher values comparing to the other eight isolates (p<0.05). For the total nitrogen, plants inoculated with ESA 90, ESA 96 and BR 5609 were not statistically different than plants supplied with NH4NO3 (p>0.05).
DiscussionAmong the typical rhizobial colonies obtained, around 53% were positive for the amplification of both symbiotic genes. The bacteria that did not showed positive results for nifH and nodC amplification are probably non-rhizobial endophytes, already observed in cowpea nodules at the Brazilian semi-arid region.35 The simultaneous amplification of symbiotic genes is a feasible tool to select the bacterial isolates which are probably rhizobia, indicating that this technique can be successfully applied for preliminary rhizobial selection.24 Among the selected bacteria, slow and fast growth characteristic were observed, which is common in soils from dry lands that harbor a large diversity of rhizobia, nodulating both crops and native species.36,37 Particularly in Brazil, a large cultural diversity of root nodule bacteria were obtained from soils at the semi-arid regions of the Pernambuco,38,39 Paraíba36 and Bahia17,38,39 states.
The 17 bacterial isolates evaluated by the API 20 NE strips metabolized several carbon sources and presented a variable enzymatic activity profile. Some isolates used 11 of the 12 carbon sources available at the API 20 NE strips, while other bacteria showed positive results only for four sources. Furthermore, some bacterial isolates showed positive enzymatic activities for six of the eight available substrates whilst other isolates were positive only for two enzymes. These results showed a large metabolic diversity of mulungu rhizobia, and possibly their adaptation in different environments.37
The clustering of bacterial isolates through the analysis of their metabolic characteristics was also related to the origin (location). The cluster 1 encompassed four isolates, among which three were isolated from Serra Talhada and only one from Caruaru. The three bacteria from Serra Talhada were classified within the Rhizobium genus. Evaluating the cluster 2, all five clustered bacteria were obtained from Caruaru and two isolates were classified within the Bradyrhizobium genus and related to B. elkanii clade. The cluster 4 also grouped two isolates from Caruaru (one Bradyrhizobium). The other clusters presented only one isolate each.
The metabolic characteristics were closely related to the differentiation of taxonomic clusters, but did not show phylogenetic relationship. This characteristic was probably observed due to the functional redundancy of the bacterial isolates, where bacteria belonging to different taxa show similar metabolic profiles, indicating the ability of bacteria from different taxonomic cluster to occupy the same ecological niche.40,41 This results indicate the importance to evaluate the diversity of rhizobia metabolic characteristics from the Brazilian Semi-Arid, to better understand their functional diversity, complementary to the molecular taxonomic assessment. Regarding the technique, the metabolic profiling of rhizobia using the API strips is widely applied in studies of new species description.42–44 This is a fast and easy test that reveals several metabolic characteristics of plant-associated bacteria,45 been applied to the characterization of rhizobial collections.
Martins at al.46 did not observed a clear distribution of Paraburkholderia isolates from Mimosa caesalpinifolia in different regions of the Brazilian semi-arid. The geographic differentiation of Bradyrhizobium isolates from nodules of field grown Chamaecrista spp. in the Semi-Arid region of the Bahia State (Northeastern, Brazil) was also not clear.47 On the other hand, the differentiation of Bradyrhizobium isolates was observed in several crop species along the Okavango Valley, from Botswana to Angola.48
For “mulungu” rhizobia in the Semi-Arid region of Brazil, there are local edaphoclimatic patterns that drives the occurrence of “mulungu” rhizobia. Menezes et al.16 evaluating “mulungu” rhizobia from soils of Juazeiro municipality (Bahia, State) from the Brazilian Semi-Arid region, observed the presence of Bradyrhizobium from the clade B. japonicum and Rhizobium within the R. tropici clade, in addition to a β-rhizobia, pointing to the occurrence of certain “mulungu” rhizobia groups according to the sampling location at the Brazilian Semi-Arid. The rhizobia found in the soils of Juazeiro by those authors are closely related to those obtained from Serra Talhada in the present study. These cities are located about 300km away. Despite the distance, the climatic characteristics of the cities are very similar regarding the temperature averages, rainfall and the plant coverage.22 Caruaru is located at 260km from Serra Talhada, but shows lower temperatures averages and higher rainfall (Table 1), changing the plant communities in the Caatinga sites,22 resulting in different soil rhizobial communities. The data obtained in this study reinforce the hypothesis that edapho-climatic conditions drive the “mulungu” rhizobial community in Brazilian Semi-Arid soils.
The bacterial isolates showed variable behavior in face of the temperature and salinity. The isolates ESA 98 and ESA 99, both B. elkanii like isolates, grew up to 45°C of incubation temperature. The isolates ESA 89 (close to B. subterraneum), ESA 97 and ESA 99 (both close to B. elkanii) highlighted due to their positive growth when inoculated in the medium supplemented with NaCl 0.51molL−1. Surprisingly, the fast growing rhizobia was not the most tolerant bacteria to temperature and salinity. Data available in the literature refers the bacteria from genus Rhizobium and other fast growers as the most tolerant isolates,13,14 probably due to their capacity to produce mucus, which provide cellular protection.49
The type strains of B. subterraneum50 and B. tropiciagri43 did not tolerate high incubation temperature grown when incubated at 37°C. Regarding the NaCl tolerance, this type of strains tolerated lower than 0.17molL−1. Generally, the bacterial isolates from semi-arid region, including the slow growing rhizobia, show higher tolerance to in vitro environmental stresses, as already observed for cowpea rhizobia regarding their tolerance to salt and temperature39 and antibiotics.51 Nevertheless, the data obtained in this study indicates that our Bradyrhizobium were more tolerant to temperature and salinity than the Bradyrhizobium evaluated by Menezes et al.,17 strengthening the hypothesis of differences among the “mulungu” rhizobia in the Brazilian Semi-Arid. Temperatures above 40°C are common in the Brazilian tropical soils, and the evaluation of the in vitro bacterial tolerance capacity to this conditions are useful to select bacterial isolates with higher resistance to harsh field conditions.
Several isolates did not solubilize the calcium phosphate in vitro. Three bacteria were positive to this characteristic, pointing out the isolate ESA 93, a Rhizobium sp. from Serra Talhada. The data obtained in this study corroborated with previous results, since low calcium phosphate solubilization are found in rhizobial collections evaluated in solid medium.52
The higher rates of auxin production were achieved in the medium with l-tryptophan. The bacteria ESA 98 and ESA 100, both Bradyrhizobium from Caruaru, produced significantly more auxin in the medium with l-try than the other bacteria. Differing from our study, the bacterial isolates evaluated by Menezes et al.17 showed that Burkholderia and Bradyrhizobium were the best auxin producers. l-try amino-acid is the main precursor of auxin and the supplementation of culture medium generally induces the auxin production by the bacterial isolates.53,54 In the medium without l-try, the bacterial isolates can produce auxin if they present other metabolic pathway for auxin production. The calcium phosphate solubilizers ESA 93 (Rhizobium sp. from Serra Talhada) and ESA 97 (Bradyrhizobium sp. from Caruaru) produced more auxin than the other bacteria in medium without l-try, showing that this isolates presents different pathways to produce auxin, a desirable characteristic regarding the plant growth promotion.12
Bradyrhizobium, Rhizobium and Paraburkholderia genera are well known for their ability to efficiently nodulate legumes in several ecological regions, including the Brazilian Semi-Arid,16,46,47,55,56 as observed in the present study. The plant inoculation experiment showed that the reference strain BR 5609 of B. elkanii, and the isolates ESA 96 and ESA 97, two B. elkanii like from Caruaru, pointed out the nodulation parameters. Other bacteria, such as the Paraburkholderia sp. ESA 96 and the Rhizobium sp. ESA 90, as well as the reference strain BR 5609, induced the same nitrogen accumulation in the shoots than the plants supplied with mineral nitrogen, indicating high symbiotic efficiency. The data reinforces the efficiency of the strain BR 5609 to E. velutina as already shown for this specie and to Erythrina falcata and E. verna. The data obtained in the present study confirm the presence of efficient “mulungu” rhizobia in the soils from semi-arid region of Brazil.16 The data also support the selection of the isolates ESA 90, ESA 96 and ESA 97, along with BR 5609, for further evaluations aiming the official recommendation for E. velutina.
Few studies had been carried out evaluating at the same time the phenotypic diversity, taxonomic classification and the symbiotic efficiency of tree species in Brazilian Semi-Arid. To our knowledge, the data obtained in the present study are the first report of the concomitantly characterization of bacteria from “mulungu” at phenotypic, molecular and symbiotic levels. In this context, the data confirm our hypothesis that the soils of different dry lands regions of Pernambuco State, under natural Caatinga regeneration harbor a diverse and efficient nitrogen-fixing bacterial community.
Further assays are needed to get a better understand of the taxonomy and the present bacteria and their potential to promote the “mulungu” growth under non-sterile conditions. In the same context, more extensive isolation experiments are needed to understand the bio-geographical patterns of rhizobial distribution in the semi-arid region of the Pernambuco State.
ConclusionsBoth α and β-rhizobia are able to occupy the E. velutina root nodules in soils of the Caatinga dry forests at the Pernambuco state (Northeastern, Brazil). The bacteria obtained showed metabolic versatility, tolerance to abiotic stresses and in vitro plant growth promotion mechanisms and the ability to induce high nitrogen accumulation in the shoots. This data set points to the potential of these bacteria to act as plant growth promoters by different ways. Furthermore, some bacteria can be selected for further studies aiming the official recommendation of the bacterial isolates for E. velutina, mainly the isolates ESA 96 and ESA 90, due to their performance in the in vivo experiment.
Conflicts of interestThe authors declare no conflicts of interest.
To the Brazilian Council for Scientific and Technological Development (CNPq–process 406327/2013-0 and 472997/2012-2) and the Brazilian Agricultural Research Corporation (Embrapa) for providing the financial support. To the Coordination of Improvement of Higher Education Personnel (CAPES) for the first to sixth authors scholarships. To the Science foundation of the Pernambuco State (FACEPE), for the scholarship to the seventh author.