In this study, phenotypic methods presented >80% agreement with the molecular identification of 59 Candida parapsilosis complex. Growth at 15% NaCl or pH 7.0 significantly reduced cfu-counts of Candida orthopsilosis, suggesting these conditions may support the development of phenotypic methods for the differentiation of the cryptic species of C. parapsilosis complex.
In 2005, Tavanti et al.1 demonstrated that the yeast known as Candida parapsilosis was actually composed by three cryptic species, and proposed the current nomenclature: C. parapsilosis sensu stricto, Candida orthopsilosis, and Candida metapsilosis. Several phenotypic assays have been used to identify C. parapsilosis species complex,2–4 but they do not accurately distinguish its cryptic species, thus requiring the use of molecular methods for differentiation. Thus, this study initially aimed to evaluate the efficacy of two phenotypical approaches to identify C. parapsilosis sensu lato, for further molecular identification of its cryptic species. Afterward, the growth kinetics of strains of C. parapsilosis sensu stricto, C. orthopsilosis, and C. metapsilosis was assessed at different salinity and pH levels to investigate phenotypic differences between these cryptic species.
The 59 yeasts included in this study were isolated from human, animal, and food samples and were presumptively identified as C. parapsilosis sensu lato, by growth on chromogenic medium, micromophological analysis on conrmeal-Tween 80 agar and biochemical analysis based on urease production, carbohydrate assimilation and fermentation and nitrogen assimilation. These microorganisms belong to the culture collection of the Specialized Medical Mycology Center of the Federal University of Ceará.
After storage, the identification of 34/59 isolates as C. parapsilosis sensu lato was confirmed by growth on chromogenic medium (CHROMagar™ Candida medium, BD™, Paris, France) and micromorphological analysis on cornmeal-Tween 80 agar, which revealed typical morphological features, i.e. branched pseudohyphae in a delicate tree-like pattern and giant cells, or features suggestive of C. parapsilosis sensu lato, such as wide elongated blastoconidia and short branched pseudohyphae.5 The other 25 isolates were re-identified by growth on chromogenic medium and automated biochemical analysis by Vitek 2™ system (bioMérieux, Jacarepaguá, Rio de Janeiro, Brazil), which showed typical biochemical profile of C. parapsilosis or biochemical features with low discrimination profile, yielding equal probability of being two or three different species, including C. parapsilosis sensu lato.
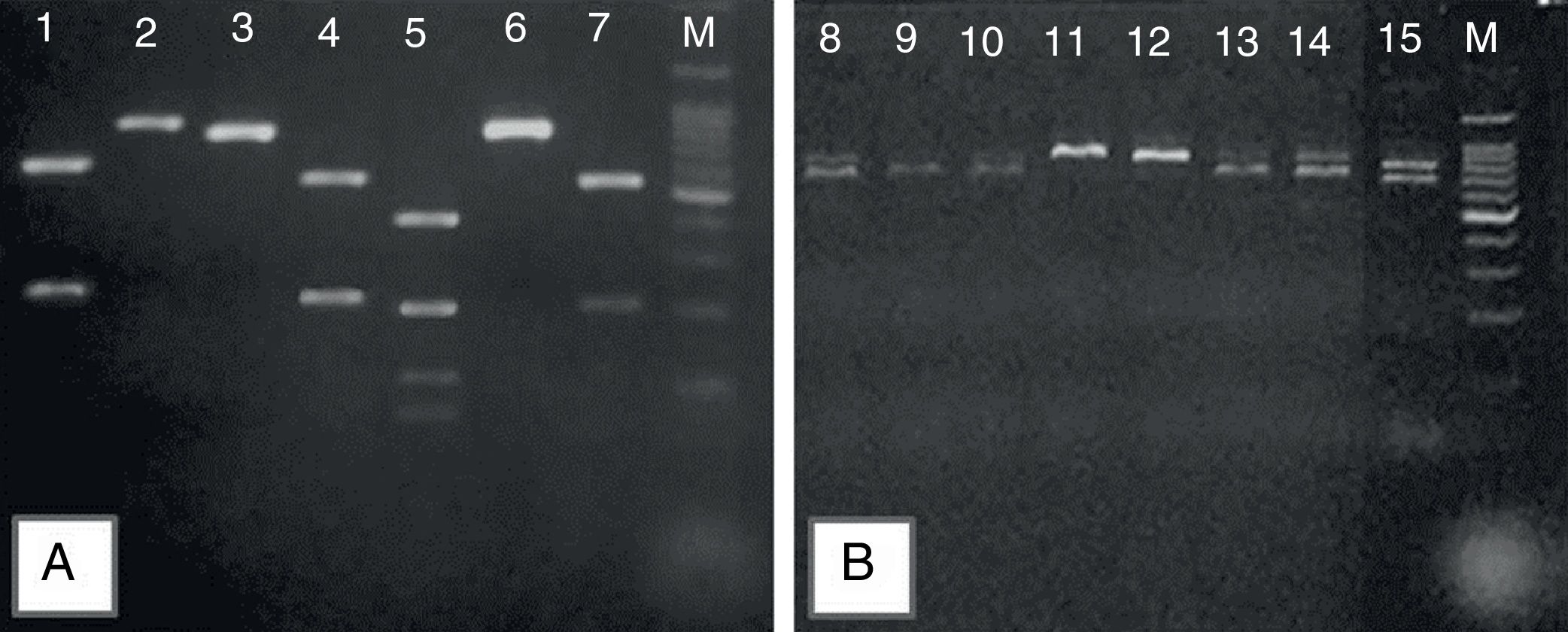
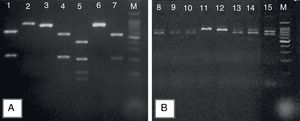
Afterwards, these 59 isolates of C. parapsilosis sensu lato were submitted to molecular identification of their cryptic species, by restriction enzyme PCR assay of the sadh gene, using the enzyme BanI, as previously described.1,6 The restriction patterns of C. parapsilosis ATCC 22019, C. orthopsilosis ATCC 96139, and a previously identified strain of C. metapsilosis6 were used to confirm identification. Six isolates with cell arrangement suggestive of C. parapsilosis sensu lato, showed unspecific band pattern, after amplification of the sadh gene. Hence, they were submitted to DNA sequencing, according to Desnos-Ollivier et al.7
Finally, six strains of each cryptic species, including the reference strains C. parapsilosis ATCC 22019 and C. orthopsilosis ATCC 96139 (Table 1), were randomly selected for the growth kinetic assays, according to Lachance et al.8 with some modifications. The strains were cultured on PDA at 35°C for 48h. Yeast inocula were prepared in sterile saline solution, with approximately 106cfu/mL, using a Neubauer chamber. Aliquots of 400μL were transferred to tubes containing 3600μL of one of the tested media. Yeasts were gown in Sabouraud broth (SAB, Difco™, Franklin Lakes, New Jersey, USA), used as cell growth control, and SAB supplemented with 5%, 10%, or 15% NaCl (w/v), or in SAB at pH 3, 5, and 7. The tubes were incubated at 35°C, and fungal growth was evaluated at 0, 24, 48, and 72h by culturing onto PDA. The plates were incubated at 35°C for 48h, and the number of cfu/mL was determined. Assays were conducted in triplicate at two different moments.
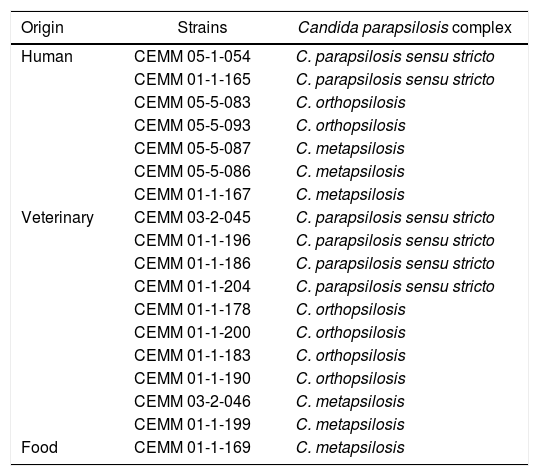
Strains of Candida parapsilosis complex selected for the study of growth kinetics.
| Origin | Strains | Candida parapsilosis complex |
|---|---|---|
| Human | CEMM 05-1-054 | C. parapsilosis sensu stricto |
| CEMM 01-1-165 | C. parapsilosis sensu stricto | |
| CEMM 05-5-083 | C. orthopsilosis | |
| CEMM 05-5-093 | C. orthopsilosis | |
| CEMM 05-5-087 | C. metapsilosis | |
| CEMM 05-5-086 | C. metapsilosis | |
| CEMM 01-1-167 | C. metapsilosis | |
| Veterinary | CEMM 03-2-045 | C. parapsilosis sensu stricto |
| CEMM 01-1-196 | C. parapsilosis sensu stricto | |
| CEMM 01-1-186 | C. parapsilosis sensu stricto | |
| CEMM 01-1-204 | C. parapsilosis sensu stricto | |
| CEMM 01-1-178 | C. orthopsilosis | |
| CEMM 01-1-200 | C. orthopsilosis | |
| CEMM 01-1-183 | C. orthopsilosis | |
| CEMM 01-1-190 | C. orthopsilosis | |
| CEMM 03-2-046 | C. metapsilosis | |
| CEMM 01-1-199 | C. metapsilosis | |
| Food | CEMM 01-1-169 | C. metapsilosis |
Growth kinetics of C. parapsilosis sensu stricto, C. orthopsilosis, and C. metapsilosis under different salinity and pH levels was analyzed, by calculating cfu-counts and average growth rate of each species: average growth rate (%)=[(number of living cells after 72h−number of living cells at time zero)/number of living cells at time zero]×100. Friedman and Kruskal–Wallis nonparametric tests were used to analyze each species, followed by Dunn's posthoc test. p-values<5% indicated significant differences.
Out of the 34 isolates reassessed by growth on chromogenic agar and micromorphology, 28 showed a typical cellular arrangement of C. parapsilosis complex, including the presence of giant cells, which were further identified as C. parapsilosis sensu stricto (n=16), C. orthopsilosis (n=8), and C. metapsilosis (n=4) (Fig. 1A), confirming the phenotypical identification of the species complex. Six isolates, on the other hand, had an arrangement similar to that of C. parapsilosis sensu lato but did not present giant cells (Table 2), showing atypical band patterns, after amplification of the sadh gene (Fig. 1B), and were identified as Candida guilliermondii by DNA sequencing. Hence, micromophological evaluation had an accuracy of 82.3% (28/34).
Representative 2% agarose gel of Candida parapsilosis sensu lato, after specific PCR reactions. (A) Products obtained after the digestion of sadh gene – PCR fragments by the BanI restriction enzyme distinguishing the cryptic species: C. parapsilosis sensu stricto (lines 1, 4, 7); C metapsilosis (line 5); and C. orthopsilosis (lines 2, 3, 6); (B) the band patterns of 716bp, obtained after sadh gene PCR reaction, suggestive of C. parapsilosis sensu lato (lines 11 and 12) and unspecific band patterns obtained for Candida guilliermondii (lines 8, 9, 10, 13, 14 and 15). Molecular marker: 1000bp.
Presumptive phenotypical identification of isolates of C. parapsilosis species complex and molecular confirmation through PCR-REA.
| Methods | Strains (n) | Chromogenic medium (n) | Phenotypical characteristics | Phenotypical identification (n) | n | Molecular identification (n) | Phenotypical identification agreement n (%) | |
|---|---|---|---|---|---|---|---|---|
| Microscopic analyses | ||||||||
| Chromogenic agar, Dalmau culture | 34 | White (17), pale pink (11) | Giant-cells, branched pseudo-hyphae and elongated blastoconidia | C. parapsilosis (28) | 28 | C. parapsilosis sensu stricto (16), C. orthopsilosis (8), C. metapsilosis (4) | 28/34 (82.3) | |
| Pale pink (6) | Wide elongated blastoconidia, with reduced filamentation | C. parapsilosis (6)b | 6 | – | ||||
| Biochemical profilea | ||||||||
| Chromogenic agar, Vitek 2 | 25 | White (3), pale pink (17) | (−) dRAFa, IMLTa, XLTa, ARBa, dCELa, ERYa, GENa, ISBEa; (v)IARAa, GRTas | (+) TyrA, dGNTa, dGATa; (v)GRTas | C. parapsilosis (20) (99%)c | 20 | C. parapsilosis sensu stricto (17), C. orthopsilosis (2), C. metapsilosis(1) | 23/25 (92.0) |
| White (1), pale pink (3) | (−) TyrA, IMLTa, LACa, XLTa, ARBa, dCELa, ERYa, GGT; (v)dXYLa, dRAFa, dGATa, GENa, GRTas | (+) dRAFa, dGNTa, GENa, ISBEa, GRTas; (v)IMLTa, dGATa (LDO) | C. famata/C. parapsilosis (4) (LDO)d | 4 | C. parapsilosis sensu stricto (1), C. orthopsilosis (2) | |||
| Pale pink (1) | (−) LysA, TyrA, IRHAa, IMLTa, NAGA1, XLTa, ARBa, dCELa; (v) dXYLa, dRAFa, IARAa, dGNTa, dGATa, GLYLa, GENa, AGLU, GRTas | (+) dRAFa, NAGa, ARG, SACa, ACEa, dGALa, dGATa, GENa, ISBEa, GRTas; (v)GLYLa, IMLTa, IARAa, dGNTa | C. famata/C. parapsilosis C. tropicalis (1) (LDO)d | 1 | – | |||
Biochemical profile (+), (−) and (v) distinct from each presumptive identification of the Vitek 2™ system. The variable biochemical profile corresponds to the profile of the strain (−) or (+) or contradictory to the given test. Biochemical tests: LysA, l-lysine arylamidase; IMLTa, l-malate; ARG, arginine; ERYa, erythritol; GLYLa, glycerol; TyrA, tyrosine arylamidase; ARBa, arbutin; dGALa, d-galactose; GENa, gentiobiose; LACa, lactose; dCELa, d-cellobiose; GGT, gamma-glutamyl-transferase; dRAFa, d-raffinose; NAGA1, PNP-N-acetyl-BD-galactosaminidase 1; dMEla, d-melibiose; ISBEa, l-sorbose; IRHAa, l-rhamnose; XLTa, xylitol; SACa, saccharose/sucrose; AGLU, alpha-glucosidase; dTREa, d-trehalose; IARAa, l-arabinose; dGATa, d-galacturonate; dXYLa, d-xylose; ACEa, acetate; GRTas, glucuronate; NAGa, N-acetyl-glucosamine; dGNTa, d-gluconate.
Micromorphologically, presented cell arrangement weakly suggestive of C. parapsilosis complex, without typical features, such as giant-cells and branched pseudohyphae in a delicate tree-like pattern, presented unspecific molecular profile and were identified as C. guilliermondii through DNA sequencing.
Among the 25 isolates identified by growth on chromogenic agar and automated biochemical analysis, 20 were confirmed as C. parapsilosis sensu lato, while five yielded inconclusive biochemical profiles, presenting equal probability of being Candida famata or C. parapsilosis (n=4) or C. famata, C. parapsilosis or Candida tropicalis (n=1) (Table 2). The molecular identification revealed that 23/25 (92%) isolates belonged to the C. parapsilosis complex (18C. parapsilosis sensu stricto, 4C. orthopsilosis and 1 C. metapsilosis) (Fig. 1A).
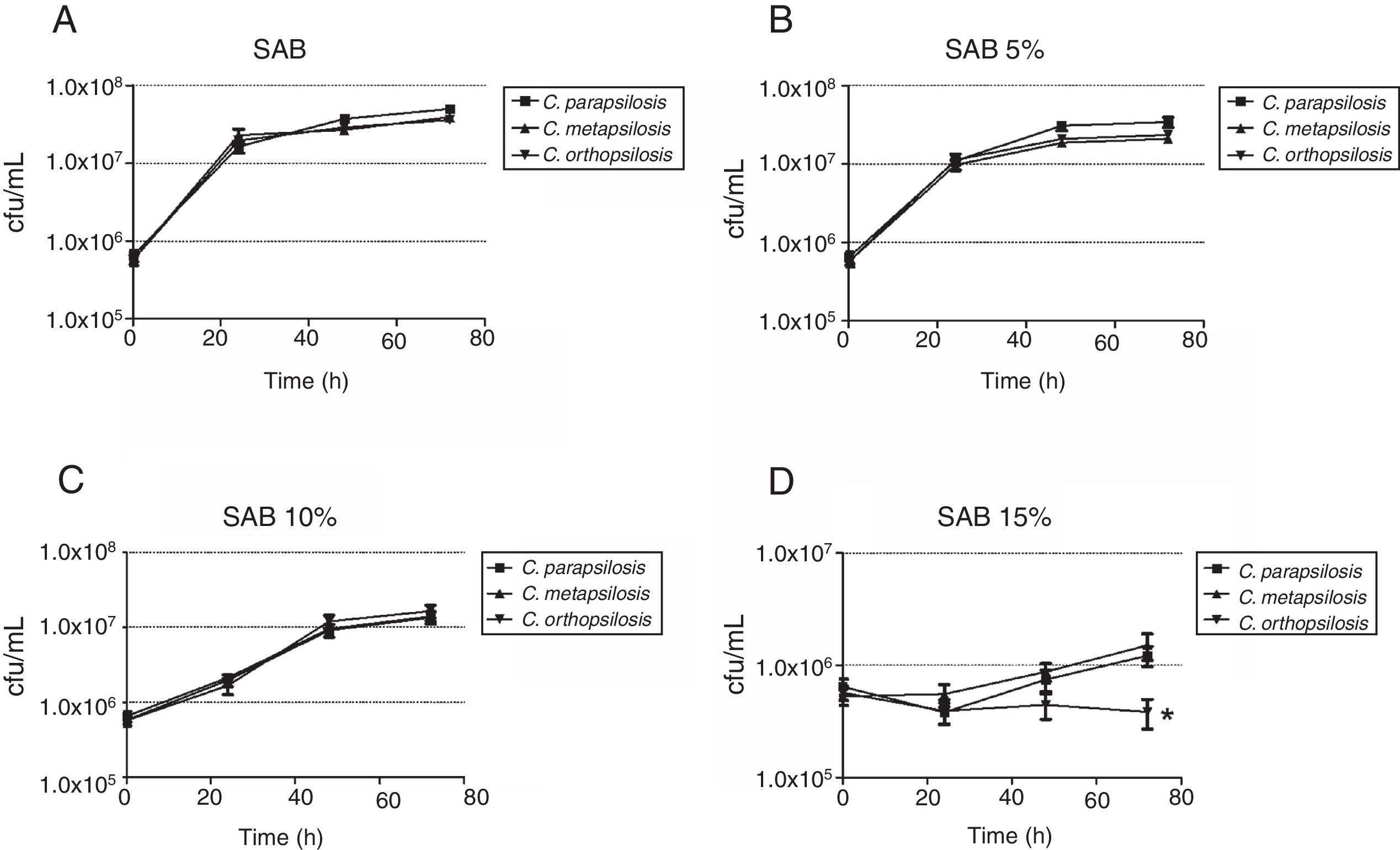
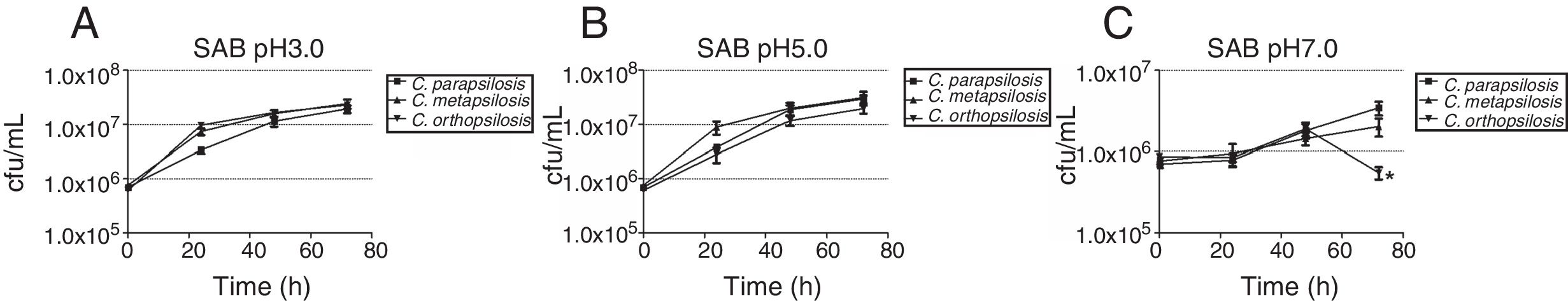
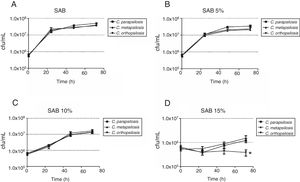
The growth kinetics at different salinities showed that maximum growth was achieved in SAB (control), and similar growth patterns were observed at 5% NaCl. The number of viable cells reduced (p<0.05) at 10% and 15% NaCl for all cryptic species, especially C. orthopsilosis, which presented significantly (p<0.05) lower average growth rate at 15% NaCl than the other two species (Fig. 2). Regarding pH levels (Fig. 3), similar growth patterns were observed at pH 3 and 5, but viable cells of the three species were significantly reduced (p<0.05) at pH 7, especially for C. orthopsilosis, which presented lower (p<0.05) average growth rate than C. metapsilosis and C. parapsilosis sensu stricto.
Growth kinetics, expressed as cfu/mL of the cryptic species of the Candida parapsilosis complex after growth up to 72h of culture in media with different salinity levels. (A) Sabouraud broth without NaCl (growth control); (B) Sabouraud broth with 5% NaCl; (C) Sabouraud broth with 10% NaCl; (D) Sabouraud broth with 15% NaCl. *Indicates statistically significant differences.
Growth kinetics, expressed as cfu/mL, of the cryptic species of the Candida parapsilosis complex, after growth up to 72h of culture in media at different pH levels. (A) Sabouraud broth pH 3.0; (B) Sabouraud broth pH 5.0; (C) Sabouraud broth pH 7.0. *Indicates statistically significant differences.
Recent studies have reported the reliability of conventional methods for the phenotypic identification of yeasts of medical importance, emphasizing that it is necessary to choose the best combination of methods to increase identification accuracy.9,10
Concerning micromorphological analysis, most strains (28/34) showed a typical morphology of C. parapsilosis sensu lato, and were correctly identified, corroborating the findings of a previous study.3 However, 6/34 isolates presented a cellular arrangement similar to that of C. parapsilosis sensu lato, but without giant cells, and were further identified as C. guilliermondii by DNA sequencing. These results show that micromorphological identification is reliable for the identification of C. parapsilosis complex, before performing the molecular analyses, when typical features are observed, with emphasis on giant cells.
Regarding the automated biochemical analysis, 80% (20/25) of the tested isolates were confirmed as C. parapsilosis sensu lato, while the remaining five isolates presented inconclusive biochemical identification. Even though the use of Vitek 2™ system has been well established in clinical microbiology laboratories for yeast identification,3,11–13 its accuracy for the identification of C. parapsilosis lato sensu, C. famata, C. tropicalis and C. guilliermondii has been considered low (approximately 90%), since low discrimination results are commonly obtained.3,9–11,14 These species have similar biochemical profiles, as observed in the present study, impairing the analysis by Vitek 2™ system, which may not differentiate between them. Alternatively, the inclusion of micromorphological analysis in the identification procedures would likely solve this issue, once conventional phenotypical methods have shown to be highly effective for the identification of C. parapsilosis sensu lato.3,10,11
The observation of epidemiological variations between the cryptic species of the complex led to the report of differences in virulence among them, with C. parapsilosis sensu stricto and C. orthopsilosis being more virulent than C. metapsilosis.15,16 Therefore, based on these epidemiological observations, phenotypic differences among the cryptic species of C. parapsilosis complex were pursued, in this study, in response to distinct growth conditions, such as salinity and pH levels. C. orthopsilosis was the least tolerant cryptic species to 15% NaCl and pH 7. These findings may explain, at least in part, the environmental distribution of the cryptic species of the C. parapsilosis complex, as they may show different phenotypic plasticity in response to distinct environmental conditions.17 Moreover, the different growth patterns of C. parapsilosis sensu stricto, C. orthopsilosis, and C. metapsilosis in hypersaline and neutral pH media may be important for further differentiation of the cryptic species.
In conclusion, micromorphological analysis and automated biochemical analysis with Vitek 2™ system are reliable approaches for phenotypic screening of C. parapsilosis complex, especially when typical morphological features and biochemical profiles are observed. Concerning growth under different salinity and pH levels, C. orthopsilosis was the least tolerant cryptic species to 15% NaCl and pH 7. These findings may partly explain the ecological differences between these cryptic species and may also be important for further identification tests, since these features may support the development of phenotypic methods for differentiating the cryptic species of C. parapsilosis complex.
Ethical approvalThis study assessed strains stored in the culture collection of the Specialized Medical Mycology Center of the Federal University of Ceará. The recovery of these strains was performed in previous studies carried out by our research group, which were approved by the Ethics Committee for the Use of Animals of the State University of Ceará.
Conflicts of interestThe authors declare no conflicts of interest.
This work was supported by grants from the National Council for Scientific and Technological Development (CNPq; Brazil; Processes 307606/2013-9, 443167/2014-1).