Anthropogenic activity, such as accidental oil spills, are typical sources of urban mangrove pollution that may affect mangrove bacterial communities as well as their mobile genetic elements. To evaluate remediation strategies, we followed over the time the effects of a petroleum hydrocarbon degrading consortium inoculated on mangrove tree Avicennia schaueriana against artificial petroleum contamination in a phytoremediation greenhouse experiment. Interestingly, despite plant protection due to the inoculation, denaturing gradient gel electrophoresis of the bacterial 16S rRNA gene fragments amplified from the total community DNA indicated that the different treatments did not significantly affect the bacterial community composition. However, while the bacterial community was rather stable, pronounced shifts were observed in the abundance of bacteria carrying plasmids. A PCR-Southern blot hybridization analysis indicated an increase in the abundance of IncP-9 catabolic plasmids. Denaturing gradient gel electrophoresis of naphthalene dioxygenase (ndo) genes amplified from cDNA (RNA) indicated the dominance of a specific ndo gene in the inoculated petroleum amendment treatment. The petroleum hydrocarbon degrading consortium characterization indicated the prevalence of bacteria assigned to Pseudomonas spp., Comamonas spp. and Ochrobactrum spp. IncP-9 plasmids were detected for the first time in Comamonas sp. and Ochrobactrum spp., which is a novelty of this study.
Mangroves are tropical and subtropical region coastal ecosystems located in transitional zones between terrestrial environments and sea and rivers.1 Several studies have demonstrated the role of the mangrove archaeal,2 bacterial3 and fungi4 communities in important biogeochemical cycles. Moreover, mangroves ecosystems have a particular and great microbial diversity. Different microorganisms inhabiting mangroves provide support for mangrove trees to survive under extreme conditions, likely by producing enzymes which are also of considerable industrial interest.5–10
In view of the current severe mangrove degradation, mainly due to the anthropogenic activity impacts, the present study is focusing on a pilot scale study for recovering these ecosystems through the adoption of bioaugmentation by using natural polycyclic aromatic hydrocarbon (PAH) degrading bacteria from the rhizosphere of mangrove plants, which might be a promising strategy to be applied in oil-polluted mangroves.11–15
The mangrove studied herein is located at Guanabara Bay, Rio de Janeiro, Brazil, near the Duque de Caxias Oil Refinery (REDUC), which has a long history of oil spill accidents and an intense raw sewage discharge into the bay.16,17 Therefore, such pollution makes the surrounding mangrove a hotspot for microbe-assisted degradation of petroleum components, such as polycyclic aromatic hydrocarbons (PAHs).18 Herein we target PAHs because they are well known to form a group of priority organic pollutants, due to the risk to the human health, high toxicity and persistence in the environment.19
Many studies have demonstrated that mobile genetic elements (MGEs), such as plasmids, play an important role in bacterial adaptation to oil biodegradation within a bacterial population, by promoting the assembly and spread of PAH catabolic gene clusters within the community.20 Among the known catabolic plasmids involved in PAH compound degradation, the narrow host range IncP-9 plasmids, which belongs to the incompatibility (Inc) plasmids group21 are of particular importance, as they often carry PAH degrading genes, such as the naphthalene dioxygenase (ndo) genes clusters.22–24 The ndo genes were target in the present study instead of others PAH degrading genes, due to their important role to initiate PAH degradation.25,26
We studied and tested a previously isolated petroleum hydrocarbon degradative consortium (PHDC) derived from the rhizosphere of the mangrove tree Avicennia schaueriana which showed a high ability for PAH removal and high abundances of IncP-9 plasmids and ndo genes.27 Herein the PHDC was characterized in order to test potential for bioremediation and application as inoculum in A. schaueriana artificially contaminated with oil in a phytoremediation greenhouse microcosm experiment. Due to the high ability for PAH removal, and high abundances of IncP-9 plasmids and ndo genes previously shown by the PHDC isolated by Gomes et al.27A. schaueriana was the mangrove tree of choice for the current microcosm experiment. This mangrove tree is typically growing in the intertidal regions of sheltered tropical and subtropical coasts,28 15–20m in size and very tolerant to high salinity.29 Microcosm experiments are often performed in the laboratory or under greenhouse conditions in order to mimic natural environment conditions as much as possible to study biodegradation pathways of different pollutants and the interactions between microorganisms and organic compounds under well controlled conditions.30
We hypothesized that the isolation of bacterial communities which have been regularly exposed to high oil concentrations might show accelerated hydrocarbon degradation, which can be explained by the proliferation of adapted bacteria involved in biodegradation pathways.31 Therefore, we expected that the PHDC inoculation to A. schaueriana grown in sediment artificially contaminated with oil, in comparison to treatments without PHDC would: (i) protect plants against deleterious oil effects, (ii) cause changes in the composition of bacterial populations, and (iii) increase the abundance of plasmids potentially carrying catabolic genes, such as ndo genes, due to the proliferation of indigenous or introduced bacteria carrying catabolic plasmids such as IncP-9 plasmids, or their spread through horizontal gene transfer.
In order to prove our hypothesis, the assessment of changes in the bacterial communities over time and the detection of functional genes and plasmids involved in PAH degradation, denaturing gradient gel electrophoresis (DGGE) analysis of bacterial 16S rRNA gene and ndo fragments, detection of ndo gene and IncP-1, IncP-7 and IncP-9 by polymerase chain reaction (PCR) and Southern blot hybridization (SBH) were performed.
Materials and methodsPetroleum hydrocarbon degrading consortium (PHDC) bioaugmentation inoculum sourceTree sampling, rhizosphere processing and PHDC enrichment were performed according to Gomes et al.27A. schaueriana was selected for the phytoremediation microcosm experiment based on previous gas chromatography (GC)-based data, which indicated a significant decrease in the total polycyclic hydrocarbon (PH) concentrations (129.6±62mgL−1) in comparison with the control flasks (301±109.3mgL−1) (p<0.05). This PHDC also showed a high abundance of naphthalene dioxygenase (ndo) genes genotypes and plasmids belonging to the incompatibility (Inc) group.27 In this study, the A. schaueriana rhizosphere PHDC were further characterized, as described below, and were used as the bioaugmentation inoculum in the phytoremediation microcosm experiment.
Petroleum hydrocarbon degrading consortium (PHDC) bacteria characterizationA cultivation-based approach combined with molecular characterization was performed, in order to characterize the bacterial community present in the inoculum, previously used in Gomes et al.,27 applied in the microcosm experiment.
Bacterial strains were isolated from the enriched inoculum applied in the microcosm experiment described below. First, 4 replicates of the inoculum were incubated in a liquid Minimum Salt Medium (MSM)32 with the addition of 1% of Arabian Light crude oil (cordially provided by Petrobras S.A.) as the only carbon source and maintained at room temperature under constant shaking at 150rpm for a total period of 20 days. After that, in order to obtain the isolates, the culture was plated in a solid MSM with crude oil vapor as the sole carbon source by dropping oil on filter paper placed on the lid of Petri dishes. The incubation time varied between 5, 15 and 20 days at 30°C. The genomic DNA was extracted33 from 80 isolates (20 from each replicate). The bacterial 16S rRNA gene was amplified in a PCR reaction by using the F-27 e R-1492 primers, generating a product of 1450bp in size.34
The PCR products were purified by using the MinElute PCR Purification Kit from QIAGEN (QIAGEN GmbH, Germany) and 100μM of the primer U8-27′ (AGA GTT TGA TCA TGG CTC AG) (Kornelia Smalla, personal communication) were added to perform the sequencing reaction. The sequencing was performed by the German Company IIT Biotech in Bielefeld, Germany.
Primers were trimmed away and chimeric sequences were removed de novo with uChime in uSearch 5.2.32.35 The sequences were compared using BLASTN (http://www.ncbi.nlm.nih.gov/BLAST) and RDP (http://rdp.cme.msu.edu/).
Plasmid DNA extraction from bacterial strains and detection of incompatibility (Inc) plasmid groups and naphthalene dioxygenase (ndo) genesBacterial strains were cultivated overnight on a MSM with oil distributed on Petri dishes covers as the only carbon source, at 28°C. Subsequently, each isolate was harvested and the plasmid DNA was obtained by phenol-chloroform extraction of the potassium acetate fraction.36 The quality of the plasmid DNA was verified under a UV light after agarose gel electrophoresis (1%, w/v) stained with ethidium bromide. Plasmid DNA from isolates was screened for the presence of ndo genes18 and of Inc plasmids belonging to the following groups: IncP-1,37 IncP-7,38 and IncP-9.20 Plasmid DNA was used for dot blot hybridization as previously described by Gomes et al.18
Experimental phytoremediation microcosm designYoung, six months old, A. schaueriana trees previously planted at the Jequiá mangrove forest located in Guanabara Bay, near the Duque de Caxias Oil Refinery (REDUC) (Rio de Janeiro, Brazil) were carefully removed along with sediment surrounding the roots and replanted into plant pots filled with 500g of sediment each. The sampling site characteristics have been previously described by Gomes et al.18 The artificial oil contamination was performed by covering the plant-mangrove sediment microcosms with 1mL of crude oil emulsified in 9mL of seawater. The following four treatments, each with four replicates, were carried out: treatment 1: plant-mangrove microcosms without addition of oil or inoculum; treatment 2: plant-mangrove microcosms inoculated with 109CFUmL−1 of the total isolated strain from the inoculum (PHDC) (pellets were obtained after centrifugation in 0.85% NaCl for 10min at 5000×g); treatment 3: plant-mangrove microcosms artificially contaminated with 1mL of emulsified crude oil in 10mL of sea water inoculated with 109CFUmL−1 inoculum (PHDC); treatment 4: plant-mangrove microcosms artificially contaminated with 1mL of emulsified crude oil in 10mL of sea water. To expose the microcosms to the high and low tide conditions, as in natural mangroves, all microcosms were partially flooded with seawater for 6h every day. Fresh seawater was directly collected from the Jequiá mangrove. At the end of the flooding period, seawater was leached out by gravitation.
Inoculum preparation and inoculationThe oil-degrading consortium was activated by cultivation in liquid MSM32 supplemented with crude oil (1%, v/v) for 5 days. The biomass production for the plant-mangrove microcosm inoculation was performed by spreading 0.1mL of the pre-activated degrading consortium onto MSM agar plates. Crude oil vapor was used as the sole carbon source by dropping oil on filter paper placed on the lid of Petri dishes. The microbial cells were harvested from the plates by washing with 0.85% saline after 3 days of incubation. The microbial suspension was centrifuged and re-suspended in sterile saline up to 3 times for removal of residual oil compounds. The cell suspension was standardized by optical density at 600nm to achieve a cell density of 109cellsmL−1. Due to the fact that mangrove roots are very sensitive to desiccation and physical damage, the plant-mangrove microcosm inoculation was performed by adding 2mL of cell suspension per each 100g of sediment above the sediment near the base of each plant. The same volume of sterile saline was added to non-inoculated soils to standardize the final moisture content of each microcosm.
Sampling and sample processingThe microcosm experiment lasted 52 days. Samples were taken just after the inoculation (day 0), and at 7, 21 and 52 days post-inoculation, corresponding to T0, T1, T2 and T3, respectively. Four entire microcosms were analyzed per treatment. The rhizosphere sample consisted of the total root system with tightly adhering sediment of four individual plants. Each rhizosphere sample was cut and thoroughly mixed prior to microbial cell detachment. Extraction of microbial cells from the roots was performed by gently shaking (100rpm) 5g of homogenized root samples in Erlenmeyer flasks containing 5g of glass beads (diameter of ∼4mm) and 45mL of extraction solution containing Tween 80 (0.1%) and sodium pyrophosphate (0.1%) for 30min. After the microbial cells were dislodged from the sediment matrices, 10mL from the supernatant of each sample were centrifuged and the pellets were re-suspended in ethanol (PA) up to a final volume of 2mL and frozen at −60°C until processed for DNA/RNA extraction.39 Only 1mL from this suspension was used for nucleic acid extraction (amount of microbial cell corresponding to 0.5g of fresh sediment).
Chlorophyll and total carotenoid pigments determinationsTo determine the stress caused in the plants due to the hydrocarbon contamination, we measured chlorophyll a and b and total carotenoid concentrations. Circular discs were cut with a 6.36mm diameter cork bore (approx. 127mm2) from both leaves of the second and third nodes counting from the top down. Each leaf disc, weighing approximately 50mg (wet weight), was placed in a vial containing 6mL of dimethyl sulfoxide (DMSO) and photosynthetic pigments were extracted without grinding at 60°C for 14h in the dark.40 Thereafter, the organic extracts were stored at 10°C and 3mL of DMSO were added to each leaf disc, maintained at 60°C for 4h for further extracting any remained pigments. These nearly colorless extracts were poured into the first ones. Chlorophylls (a+b) and total carotenoids (carotenes plus xanthophylls) were quantified spectrophotometrically according to Wellburn41 after subtracting the turbidity at 750nm.
Total community (TC)-DNA and cDNA (RNA) extraction from microcosm soil samplesTC-DNA was extracted from 0.5g of microcosm soil samples using the FastDNA® Spin Kit for Soil (Bio101, Qbiogene, Carlsbad, CA, USA) after a harsh lysis step with the FastPrep FP120 bead beating system for cell lysis. DNA was purified by GENECLEAN Spin Kit (Qbiogene), and the yield and quality were verified by electrophoresis on 1% (w/v) agarose gels under UV light after staining with ethidium bromide. The extracted TC-DNA was used for PCR-Southern blot hybridization (SBH) analysis, quantitative real-time PCR (qPCR) and DGGE fingerprints. The absence of PCR-inhibiting substances was verified by PCR amplification of bacterial 16S rRNA gene ragments from TC-DNA using the F27 and R1494 primers as previously described by Heuer et al.42 The cDNA was extracted according to Costa et al.43
Bacterial 16S rRNA gene PCR amplification and quantificationBacterial 16S rRNA gene PCR amplification was performed as previously described by Heuer et al.42 (product size of 1506bp) from the TC-DNA of microcosm soil samples and the genomic DNA from isolated bacterial strains. PCR products were checked after electrophoresis in 1% agarose gel stained with ethidium bromide under UV light in comparison with the 1-kb gene-ruler TM DNA ladder (Fermentas, St. Leon-Rot, Germany). Quantitative PCR (qPCR) targeting the bacterial 16S rRNA gene from the TC-DNA was performed with the TaqMan system as described by Suzuki et al.44 The bacterial 16S rRNA gene qPCR standard was obtained through a dilution series (10−3–10−7) of the cloned bacterial 16S rRNA gene amplicons (1467bp) from Escherichia coli.
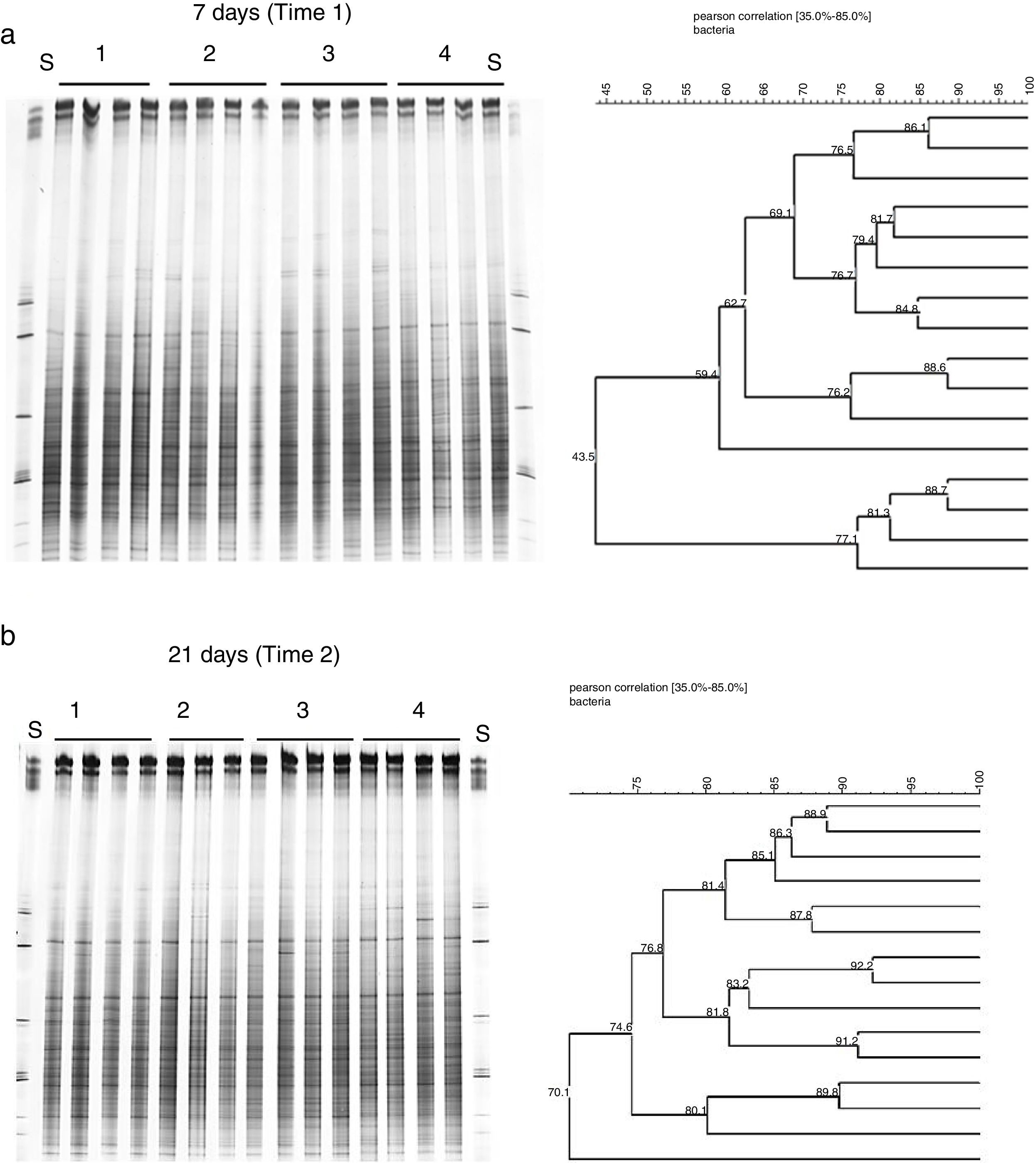
DGGE analysis of bacterial 16S rRNA gene fragments from the TC-DNA of microcosm soil samplesBacterial 16S rRNA gene fragments amplified from TC-DNA were analyzed by DGGE as described by Ding et al.45 Comparison of silver-stained DGGE profiles and band intensities was performed by means of the GelCompar II 5.6 software (Applied Maths, Sint-Martens-Latem, Belgium). A cluster analysis based on the Pearson similarity matrix was constructed by the Unweighted Pair Group Method with Arithmetic Mean (UPGMA).46 In order to evaluate slight differences in the bacterial community fingerprints visualized by DGGE, a statistical test analysis was applied by using a permutation test based on the pairwise similarity measures.47
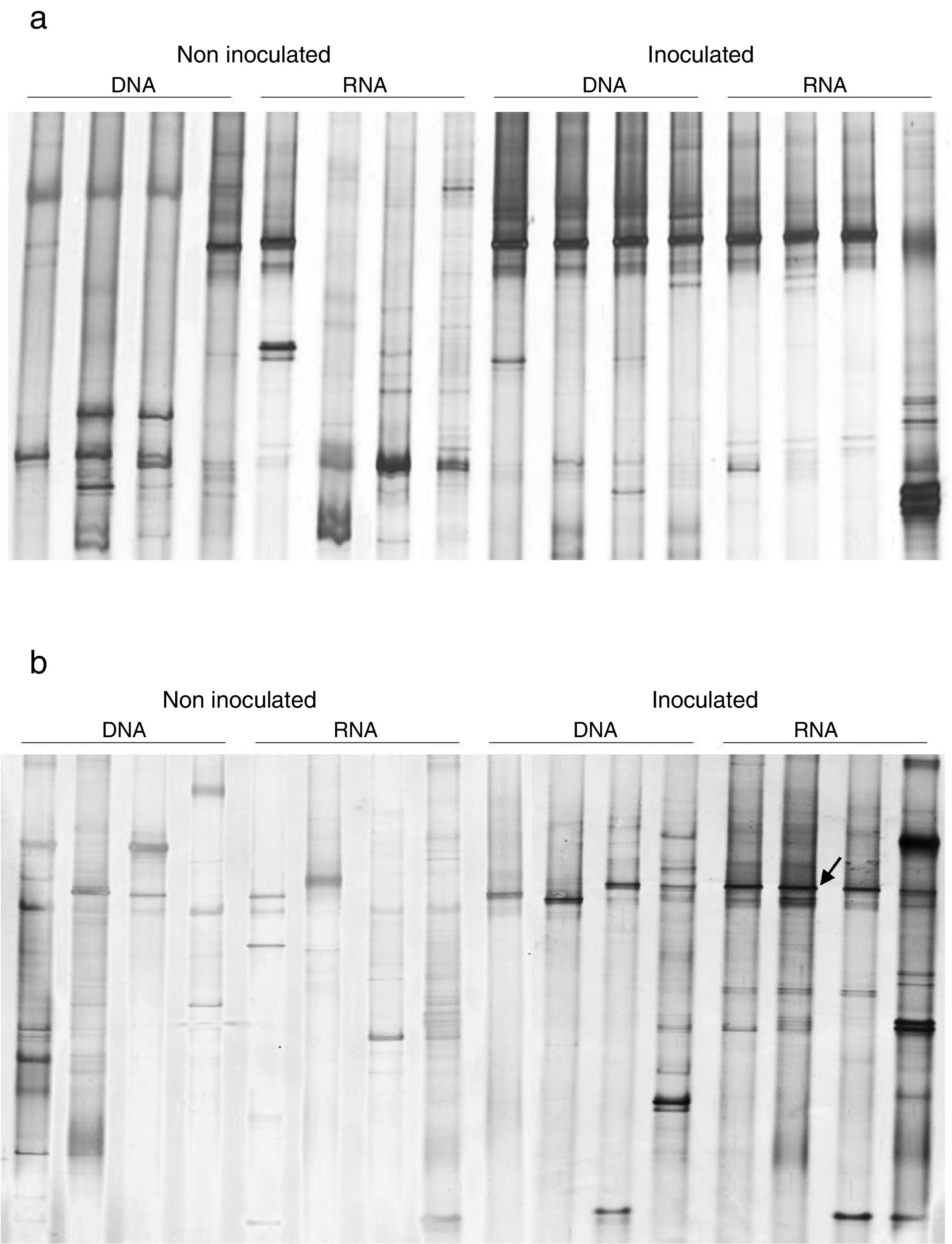
Naphthalene dioxygenase (ndo) gene-based DGGE analysisPCR amplification of ndo gene fragments from the TC-DNA and cDNA of the four microcosm treatments at two different sampling times were performed for DGGE analyses.18 PCR products were checked on 1% agarose gels in order to verify the correct band size. The ndo genes were subsequently analyzed by DGGE as described by Gomes et al.18
Southern blot-PCR based detection of incompatibility (Inc) plasmids: IncP-1, IncP-7 and IncP-9 plasmids from the TC-DNA of microcosm soil samplesThe Southern blot-PCR based detection of IncP-1,37 IncP-7,38 and IncP-920 from the TC-DNA of the four microcosm treatments at two different sampling times was performed according to Dealtry et al.48
ResultsIdentification of bacterial isolates from the PHDC inoculum based on the partial sequence of the bacterial 16S rRNA geneFrom the initial 80 isolates, a screening based on phenotypes and biochemical tests: colony morphology, catalase and oxidase reaction,49 and Gram staining test,50 led to the selection of 35 isolates for bacterial 16S rRNA partial gene sequencing. Based on the partial sequences of the 16S rRNA gene, the bacterial strains were identified as Pseudomonas spp. (20), Ochrobactrum spp. (6), Comamonas spp. (5), Achromobacter spp. (2), Bacillus sp. (1), and Pseudoacidovorax sp. (1). The bacterial 16S rRNA amplicon sequences have GenBank accession numbers KX061935–KX061969.
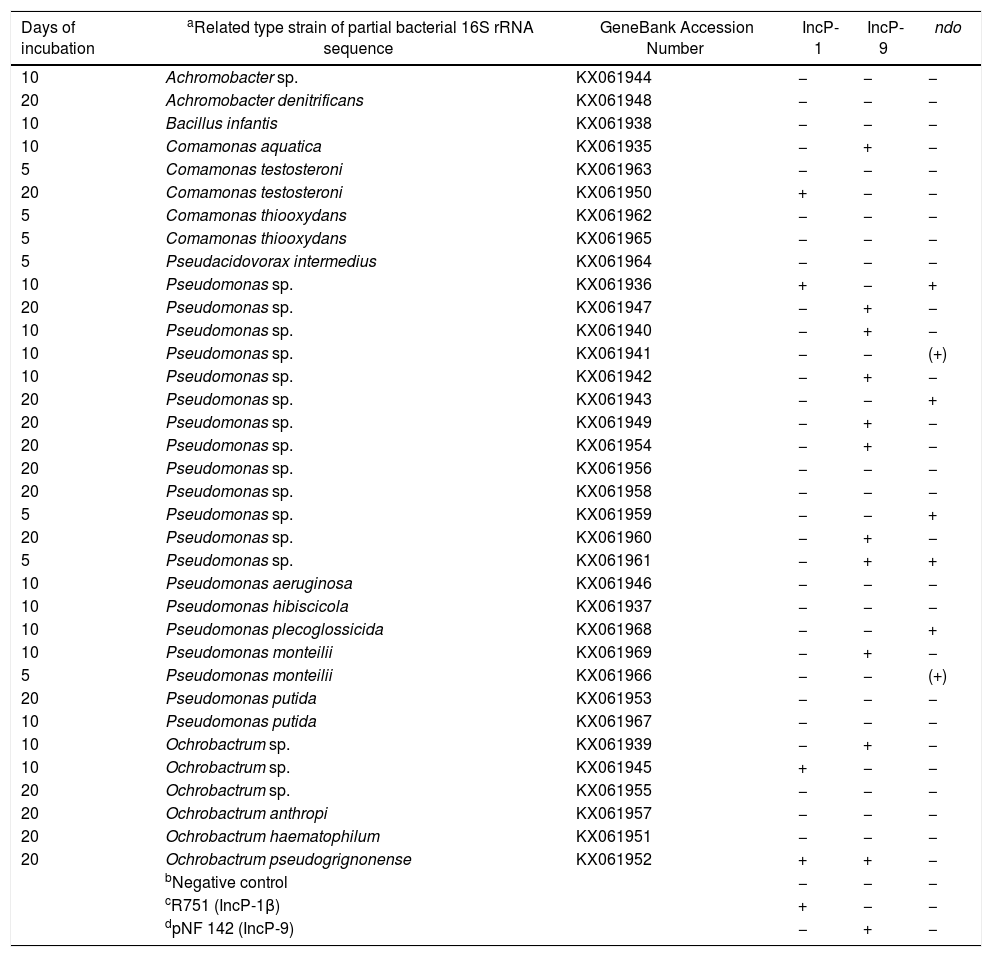
Detection of incompatibility (Inc) plasmids and naphthalene dioxygenase (ndo) genes from the plasmid DNA of PHDC bacterial isolatesThe presence of plasmids and ndo genes were confirmed by dot blot hybridization analysis performed with the PCR products with predicted band sizes of each of the amplicon targets. PCR amplicons and Southern blot hybridization (SBH) signals of the predicted sizes corresponding to IncP-1 plasmids were obtained for five isolates. Most of the isolates (11), showed PCR amplicons and SBH signals corresponding to IncP-9 plasmids (Supplementary S1), while six showed signals indicating the presence of ndo genes (Supplementary S2). Only one isolate showed the presence of IncP-9 plasmid and ndo gene at same time (Table 1).
Identification of bacteria isolated from the petroleum hydrocarbon degradative consortium (PHDC), based on bacterial 16S rRNA gene partial sequence and screening for the detection of incompatibility (Inc) plasmids and naphthalene dioxygenase (ndo) genes trough dot blot hybridization.
| Days of incubation | aRelated type strain of partial bacterial 16S rRNA sequence | GeneBank Accession Number | IncP-1 | IncP-9 | ndo |
|---|---|---|---|---|---|
| 10 | Achromobacter sp. | KX061944 | − | − | − |
| 20 | Achromobacter denitrificans | KX061948 | − | − | − |
| 10 | Bacillus infantis | KX061938 | − | − | − |
| 10 | Comamonas aquatica | KX061935 | − | + | − |
| 5 | Comamonas testosteroni | KX061963 | − | − | − |
| 20 | Comamonas testosteroni | KX061950 | + | − | − |
| 5 | Comamonas thiooxydans | KX061962 | − | − | − |
| 5 | Comamonas thiooxydans | KX061965 | − | − | − |
| 5 | Pseudacidovorax intermedius | KX061964 | − | − | − |
| 10 | Pseudomonas sp. | KX061936 | + | − | + |
| 20 | Pseudomonas sp. | KX061947 | − | + | − |
| 10 | Pseudomonas sp. | KX061940 | − | + | − |
| 10 | Pseudomonas sp. | KX061941 | − | − | (+) |
| 10 | Pseudomonas sp. | KX061942 | − | + | − |
| 20 | Pseudomonas sp. | KX061943 | − | − | + |
| 20 | Pseudomonas sp. | KX061949 | − | + | − |
| 20 | Pseudomonas sp. | KX061954 | − | + | − |
| 20 | Pseudomonas sp. | KX061956 | − | − | − |
| 20 | Pseudomonas sp. | KX061958 | − | − | − |
| 5 | Pseudomonas sp. | KX061959 | − | − | + |
| 20 | Pseudomonas sp. | KX061960 | − | + | − |
| 5 | Pseudomonas sp. | KX061961 | − | + | + |
| 10 | Pseudomonas aeruginosa | KX061946 | − | − | − |
| 10 | Pseudomonas hibiscicola | KX061937 | − | − | − |
| 10 | Pseudomonas plecoglossicida | KX061968 | − | − | + |
| 10 | Pseudomonas monteilii | KX061969 | − | + | − |
| 5 | Pseudomonas monteilii | KX061966 | − | − | (+) |
| 20 | Pseudomonas putida | KX061953 | − | − | − |
| 10 | Pseudomonas putida | KX061967 | − | − | − |
| 10 | Ochrobactrum sp. | KX061939 | − | + | − |
| 10 | Ochrobactrum sp. | KX061945 | + | − | − |
| 20 | Ochrobactrum sp. | KX061955 | − | − | − |
| 20 | Ochrobactrum anthropi | KX061957 | − | − | − |
| 20 | Ochrobactrum haematophilum | KX061951 | − | − | − |
| 20 | Ochrobactrum pseudogrignonense | KX061952 | + | + | − |
| bNegative control | − | − | − | ||
| cR751 (IncP-1β) | + | − | − | ||
| dpNF 142 (IncP-9) | − | + | − |
All sequences of identified bacterial species showed 100% of similarity with the type strains compared in the Ribosomal Database Project (RDP) data base.
Negative control was represented by a randomly isolated bacterium from a soil contaminated with oil, which previously PCR-Southern blot hybridization analyzes showed the absence of IncP-1, IncP-7 and IncP-9 plasmids.
R751 (IncP-1β) was obtained from E. coli CM544 and cordially provided by the group of Sørensen SJ.37
pNF 142 (IncP-9) was isolated from a creosote-contaminated soil.39
(+) possible variation of the ndo detected.
In order to evaluate the potential of the inoculated PHDC to protect A. schaueriana mangrove plants against the phytotoxicity induced by artificial contamination with petroleum, symptoms such as the numbers of leaves with blight lesions and plant length at the initial time point (T0) and at the end of the experiment (T3) were measured and compared. The results indicated that none of the analyzed leaves displayed blight lesions, and only weak symptoms in five leaves were observed in one replicate (3.1) of the inoculated plants artificially contaminated with oil at the end of the experiment (T3) in the control (1) and in inoculated plants without oil (2). On the other hand, all plants artificially contaminated with oil (4) displayed several leaves with blight lesions: six, three, nine and four leaves respectively, from 4.1, 4.2, 4.3 and 4.4 at the end of the experiment (T3) (Fig. 1).
Effects of PAH contamination on A. schaueriana mangrove tree stems and blight lesions after 21 days of contamination with crude oil. (A) controls, plants without inoculum and oil; (B) inoculated plants; (C) inoculated plants artificially contaminated with 1mL of emulsified crude oil in 10mL of sea water; (D) plant artificially contaminated with 1mL of emulsified crude oil in 10mL of sea water.
In order to evaluate A. schaueriana stress caused by the artificial addition of petroleum, pigment extractions from the initial time point (T0) and the end of the experiment (T3) were performed. The results of these pigments, total chlorophyll (a+b) and total carotenoids, indicate that, except for the control plants (1), all other treatments, 2, 3 and 4, respectively – inoculated plants, inoculated plants artificially contaminated with oil, and plants artificially contaminated with oil – resulted in a decrease of chlorophyll (a+b) levels and, to a lower degree, in a decrease of total carotenoids. The occurrence of plant stress is indicated by the ratio of chlorophyll divided by the total carotenoid concentrations, which, in the present study, decreased significantly from the ratio of 6.2mg/m2 (control) to 5.2mg/m2 in the other treatments (2; 3; 4) at the last sampling timepoint.
Determination of bacterial 16S rRNA gene copies by qPCR from the TC-DNA of microcosm soil samplesIn order to estimate the bacterial abundance of the mangrove microcosm samples, the 16S rRNA gene copies were determined by qPCR from soil TC-DNA extracts. All samples (Table 2) showed a high abundance of bacterial populations ranging from 108 to 109 bacterial 16S rRNA gene copy numbers per gram of microcosm soil samples with no significant differences between the treatments (Tukey's test, p<0.05).
Bacterial densities and PCR-Southern blot hybridization detection of plasmid replicon-specific sequences belonging to the IncP-1, IncP-7 and IncP-9 groups.
| Sample | Description of samples | IncP-1 | IncP-7 | IncP-9 | 16S rRNA gene log10/g |
|---|---|---|---|---|---|
| 1 | Treatment 1.1 Time 1 | ++ | − | − | |
| 2 | Treatment 1.2 Time 1 | ++ | − | − | |
| 3 | Treatment 1.3 Time 1 | ++ | − | + | 9.02a |
| 4 | Treatment 1.4 Time 1 | + | − | − | |
| 5 | Treatment 1.1 Time 2 | ++ | − | − | |
| 6 | Treatment 1.2 Time 2 | + | − | − | |
| 7 | Treatment 1.3 Time 2 | − | − | − | 8.79a |
| 8 | Treatment 1.4 Time 2 | ++ | − | − | |
| 9 | Treatment 2.1 Time 1 | ++ | − | ++ | |
| 10 | Treatment 2.2 Time 1 | ++ | − | + | |
| 11 | Treatment 2.3 Time 1 | − | − | ++ | 8.85a |
| 12 | Treatment 2.4 Time 1 | +++ | − | ++ | |
| 13 | Treatment 2.1 Time 2 | ++ | − | + | |
| 14 | Treatment 2.2 Time 2 | ++ | − | + | |
| 15 | Treatment 2.3 Time 2 | + | − | + | 9.07a |
| 16 | Treatment 2.4 Time 2 | + | − | − | |
| 17 | Treatment 3.1 Time 1 | +++ | − | +++ | |
| 18 | Treatment 3.2 Time 1 | ++ | − | +++ | |
| 19 | Treatment 3.3 Time 1 | ++ | − | +++ | 9.07a |
| 20 | Treatment 3. Time 1 | ++ | − | ++ | |
| 21 | Treatment 3.1 Time 2 | − | − | + | |
| 22 | Treatment 3.2 Time 2 | ++ | − | + | |
| 23 | Treatment 3.3 Time 2 | ++ | − | ++ | 9.18a |
| 24 | Treatment 3.4 Time 2 | − | − | ++ | |
| 25 | Treatment 4.1 Time 1 | + | − | − | |
| 26 | Treatment 4.2 Time 1 | − | − | + | |
| 27 | Treatment 4.3 Time 1 | ++ | − | − | 8.69a |
| 28 | Treatment 4.4 Time 1 | − | − | − | |
| 29 | Treatment 4.1 Time 2 | ++ | − | − | |
| 30 | Treatment 4.2 Time 2 | + | − | + | |
| 31 | Treatment 4.3 Time 2 | ++ | − | − | 8.9a |
| 32 | Treatment 4.4 Time 2 | + | − | + | |
| 33 | Inoculum 1 | − | − | +++ | |
| 34 | Inoculum 2 | − | − | +++ | |
| 35 | Inoculum 3 | − | − | +++ | |
| bNegative control | − | − | − | ||
| cR751 (IncP-1β) | +++ | ||||
| dpCAR1 (IncP-7) | +++ | ||||
| epNF 142 (IncP-9) | +++ |
Hybridization signal: (+++) very strong, with exposure time up to 5min; (++) strong, with exposure time up to 1h; (+) weak, with exposure time up to 3h; (−) none, with exposure time of more than 3h.
Negative control was represented by a randomly isolated bacterium from a soil contaminated with oil, which previously PCR-Southern blot hybridization analyzes showed the absence of IncP-1, IncP-7 and IncP-9 plasmids.
R751 (IncP-1β) was obtained from E. coli CM544 and cordially provided by the group of Sørensen SJ.37
pCAR1 (IncP-7) was obtained from Pseudomonas resinovorans CA10 and cordially provided by the group of Izmalkova TY.38
pNF 142 (IncP-9) was isolated from a creosote-contaminated soil.39
Time 1=7 days after inoculation; Time 2=21 days after inoculation.
Treatment 1.1; 1.2; 1.3 and 1.4: control, plants without inoculum and oil; treatment 2.1; 2.2; 2.3 and 2.4: inoculated plants; treatment 3.1; 3.2; 3.3 and 3.4: inoculated plants artificially contaminated with 1mL of emulsified crude oil in 10mL of sea water; treatment 4.1; 4.2; 4.3 and 4.4: plant artificially contaminated with 1mL of emulsified crude oil in 10mL of sea water.
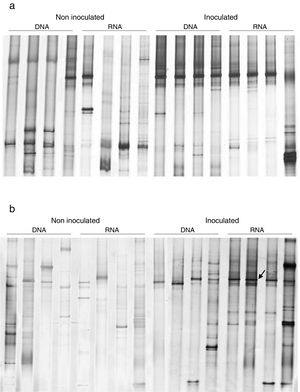
To evaluate the effect of oil and PHDC on the bacterial community composition over time, a DGGE analysis was performed of bacterial 16S rRNA genes amplified from TC-DNA extracted from the four microcosm treatments from samples taken after 7 and 21 days of growth in the greenhouse. The bacterial DGGE fingerprints (Fig. 2a and b) showed a high stability of the bacterial community over time, without detectable variation among replicates and treatments. The UPGMA cluster analysis of the DGGE patterns did not detect any significant difference between the two sampling times for the bacterial diversity between the different treatments over time (Fig. 2a and b). The permutation test analysis of total bacterial DGGE fingerprints revealed no significant differences in the composition of these communities between the different treatments and incubation times. The DGGE analysis data is only shown for the time point after seven (T1) and 21 (T2) days of inoculation in congruence with the time points analyzed by the others molecular analysis (ndo-DGGE fingerprints of DNA and cDNA genes and Southern blot hybridization of catabolic plasmids).
Comparison of the bacterial community structures by DGGE based on bacterial 16S rRNA. S: (S) Standard bacterial 16S rRNA; (1) controls, plants without inoculum and oil; (2) inoculated plants; (3) inoculated plants artificially contaminated with 1mL of emulsified crude oil in 10mL of sea water; (4) plant artificially contaminated with 1mL of emulsified crude oil in 10mL of sea water.
In this study, an ndo gene fingerprint approach18 was used to evaluate the effect of oil contamination and bioaugmentation on the diversity and expression of polycyclic aromatic hydrocarbon (PAH) degrading genes. Based on the band profiles, the comparison of ndo-DGGE fingerprints of DNA and cDNA revealed that both indigenous and introduced bacteria were able to express the genes coding for PAH degrading enzymes in the sediment rhizosphere (Fig. 3a and b). However, the appearance of dominant bands in the cDNA of inoculated treatments was observed, with the most intense signal on day 7, indicated the dominance of a specific ndo gene in the inoculated treatments, with high abundance and expression on day 7.
Southern Blot-PCR based detection of incompatibility (Inc) plasmids from soil TC-DNAFrom the TC-DNA, Southern blot hybridization (SBH) with labeled probes generated from specific PCR-amplicons from the TC-DNA were applied in order to avoid the detection of false-positive results which might occur in a conventional PCR amplification.51 In order to detect the occurrence and temporal and treatment-dependent changes in the abundance of IncP-1, IncP-7 and IncP-9 plasmids, SBH analysis were performed.
PCR amplicons and SBH signals of the predicted sizes were not obtained for IncP-7 plasmids (Table 2). However, weak, strong and very strong signals were detected for IncP-1 and IncP-9 plasmids. While SBH of IncP-1 signals were detected without variation over time and between the different treatments, SBH of IncP-9 signals increased in response to the addition of the PHDC consortium in oil polluted treatments over the time. The SBH analysis results indicated higher IncP-9 plasmid abundance in the oil treatment, displaying the strongest signals (S3) in treatment 3 with the respective replicates on day 7 (T1), decreasing IncP-9 plasmid abundance after 21 days (T2) (Table 2). Southern Blot-PCR-based detection of Inc plasmid analysis data is only shown for the time point after seven and 21 days of inoculation, due to the absence of variation from the day 21 (T2) and at the end of the experiment, after 52 days of inoculation (T3).
DiscussionThe microbial community was exposed to Arabian Light crude oil in the greenhouse microcosm experiment with or without the addition of selected indigenous petroleum hydrocarbon degrading consortium (PHDC). The microcosm experiment simulated an oil spill, and the effects of the microbial inoculation were evaluated over the time. The PHDC tested as inoculum in this phytoremediation microcosm experiment was previously shown to efficiently degrade polyaromatic hydrocarbons.27 Previous studies reported high abundances of IncP-9 plasmids and naphthalene dioxygenase (ndo) genes by Southern blot hybridization (SBH) analysis, and considerable polycyclic aromatic hydrocarbon (PAH) removal rates detected in PHDC originated from the A. schaueriana rhizosphere.27 In addition, a microarray analysis of the PHDC indicated that the majority of taxa with positive microarray signals, and most of the populations detected by the sequence analysis of dominant DGGE bands were associated to Pseudomonadales. Moreover, the bacterial 16S rRNA gene sequence analysis of bands excised from Pseudomonas DGGE profiles indicated the dominance of populations related to P. aeruginosa and P. putida, which are commonly involved in polycyclic hydrocarbons (PHs) degradation.27 However, the bacterial population present in the PHDC must be alive to be used as inoculum, and cultivation-independent TC-DNA based methods do not reflect survival. Therefore, bacterial isolates were obtained by using crude oil as selection source in the present study, in order to characterize the PHDC used as inoculum in the A. schaueriana phytoremediation microcosm experiment. As expected because the IncP-9 plasmid detected, most of the isolates were assigned to Pseudomonas spp. However, the majority of the isolates carried IncP-9 and IncP-1 plasmids and ndo genes, indicating their potential to perform the first step in PAH degradation. The isolation of PAH degrading bacteria is important for the formulation of efficient PHDC based inocula to be used in future phytoremediation applications, where the processes are dependent on microbial viability. Interestingly, some of the bacterial isolates assigned to Pseudomonas spp. (Table 1), which are well-known PAH degraders,52–56 presented ndo genes in the absence of IncP-9 plasmids, which often carry genes involved in petroleum degradation and other natural contaminants.20 We also observed the presence of ndo genes in one of Pseudomonas sp. isolate which carried only IncP-1 plasmid, generally described as vehicle of genes involved in the degradation of man-made pollutants, such as pesticides and not natural contaminants such as petroleum.24
IncP-1 plasmids are a broad host range and have been observed in a wide range of hosts, such as Achromobacter xylosoxidans, Burkholderia cepacia, Cupriavidus necator, Pseudomonas sp., Sphingomonas sp. A1, Variovorax sp.,57 while IncP-9 is well known to be a narrow host range plasmid, mainly occurring in Pseudomonas.58 However, IncP-9 was detected in bacteria from other taxonomic groups for the first time here: in Comamonas aquatica and in two different Ochrobactrum spp. species. These are very interesting findings, which may also indicate that environmental pressure over time might have fostered the adaptation of other hosts by the uptake of IncP-9 plasmids, in order to increase bacterial survival under stressful conditions.
The use of bioremediation to accelerate the rates of petroleum components degradation has many successful examples, such as the case of the Exxon Valdez oil spill in Prince William Sound59 and the oil spill in the northern Gulf of Mexico.60 The results obtained in these cases have demonstrated that the stimulation of the indigenous bacteria able to degrade hydrocarbons by adding fertilizer significantly enhanced petroleum degradation rates and shaped the function and structure of bacterial communities.61 The use of plants to enhance the bioremediation of petroleum hydrocarbons at contaminated soil sites, at a pilot-scale, demonstrated the relevant role of rhizosphere-associated microbes in the petroleum components degradation.62,63 The bioaugmentation strategy used in the present greenhouse microcosm indicated promising results and can be further explored in future field phytoremediation experiments of polluted mangrove ecosystems.
The short-term effects of oil addition in association with the PHDC inoculation were evaluated under both aspects: structure and functional level. Firstly, in order to unravel and assess the changes in the structure of bacterial communities, a culture-independent method, the PCR-DGGE fingerprint technique, which is a suitable technique to access the structure of bacterial and archaeal communities of mangrove trees,64 was applied. Pyrosequencing analysis of bacterial 16S rRNA gene amplicons already showed that most of the microbial community of mangrove trees involved in oil degradation were assigned to Bacteria, with highest abundance related to Proteobacteria,65,66 which guided our focus to the comparative analysis of bacterial communities inhabiting the rhizosphere of mangrove trees. Based on the appearance of several DGGE bands in both analyzed sampling times, bacterial PHDC fingerprints indicated a high complexity of different bacterial populations. These results are indications of the adaptation of a high number of populations able to survive in the presence of PHs. These findings are in accordance with the results reported by Gomes et al.,27 which revealed a higher number of bacterial populations adapted to growth on PH, inhabiting the rhizosphere, in comparison with the complexity found in the bulk sediment.
Analysis of photosynthetic pigments (chlorophyll a, chlorophyll b and carotenoids) are often performed to indicate plant stress.67 In comparison with the control plants at the end of the experiment (T3), the chlorophyll/carotenoid ratio (total chlorophyll over total carotenoid) showed increased plant stress for all other evaluated treatments indicating plant metabolism alterations induced by the inoculated microorganisms. On the one hand, pigment analysis did not indicate a decrease of plant stress induced by oil due to the PHDC inoculation. On the other hand, plant symptoms observed in leaves indicate protection against oil-induced phytotoxicity.68 Our results thus demonstrate that bacterial communities able to partially or totally degrade PHs inoculated in the greenhouse microcosm plants could improve A. schaueriana health, protecting this species against the phytotoxicity induced by the artificial contamination with petroleum. Inoculated plants showed only weak symptoms in five leaves in one replicate, while the non-inoculated plants showed several leaves with blight lesions (Fig. 1), indicating the protective effect of the inoculation with PAH-degrading bacteria in the presence of oil. The absence of significant differences in plant length measured at the end of the experiment (T3) (data not shown) between the non-inoculated and inoculated plants can be explained by the use of trees originated from a polluted urban mangrove, which likely carried oil-adapted bacterial communities, enriched catabolic plasmids and PAH degrading genes.27,31 The frequent addition of petroleum to a microcosm tends to affect the microbial communities present in this environment, which also occurs in situ, in different polluted mangroves.23 The selection of PAH-degrading bacteria occurs in order to maintain the health and balance of this ecosystem. The shifts in the structure of pre-adapted bacterial communities to long-term oil pollution tending toward lesser diversity, but more specialized and adapted microbial communities, have been previously reported,45,69 in accordance with the bacterial community stability shown in both treatments, as revealed by the DGGE analysis herein.
Secondly, in order to unravel and assess functional changes in bacterial communities caused by the short-term effects of oil addition in association with the PHDC inoculation, analysis of mobile genetic elements (MGEs) of interest were performed. Interestingly, while the structure of bacterial community was rather stable, pronounced shifts occurred in the MGEs over the time, indicating that the adaptation process in the mangrove is probably mainly driven by the proliferation of bacteria carrying plasmids with degradative genes and horizontal transfer of catabolic plasmids, e.g. encoding PAH degrading genetic determinants, rather than by changes in bacterial community composition due to the selection for PAH-degrading bacteria already present in the environment.24
Although SBH does not provide quantitative information on specific gene copy numbers per gram of material as does qPCR, changes in the abundances of the different plasmid groups can be indicated and are often confirmed by amplicon pyrosequencing, as demonstrated in other studies involving the biodegradation of organic pollutants.24 On the basis of the exposure time of the identical blots hybridized with digoxigenin-labeled probes generated from reference plasmids (S1), it was shown that the bacteria carrying IncP-9 plasmids were highly abundant in treatment 3, which received both oil and the PHDC inoculum. The comparison between the bacterial 16S rRNA gene copy numbers by qPCR indicated that these differences were not due to differences in bacterial abundance among the samples. Several other studies have demonstrated the role of IncP-9 plasmids in the adaptation of the bacterial communities to PAH contamination in situ.58,70 The high abundance of the IncP-9 plasmids observed in the present study confirmed previously reported findings20 which indicate that IncP-9 plasmids often carry genes involved in the degradation of natural contaminants, e.g. ndo gene products responsible for different PAH degradation steps.18 The exposure of mangrove bacteria to PAH for a longer period of time might have fostered adaptations to rapidly changing environmental conditions via horizontally acquired MGEs.31 No treatment-dependent changes in the abundance of IncP-1 plasmid carrying populations were observed, which is not surprising, since it is well known that this plasmid group often encodes genes involved in man-made pollutants, e.g. pesticides20 and not natural organic pollutants like petroleum.
It is well known that the important multicomponent enzyme system involved in PAH degradation, ndo genes, present a huge genetic variability which complicates their detection in environmental samples. However, in order to overcome this limitation, primers which target the ndo alpha subunits belonging to the main clade of group III, comprising several ndo genes, including the nahAc and phnAc genes, were applied.18 PCR-based primer systems targeting a broad range of ndo genes were applied, and it was shown that the enrichment with microbial PAH genes indeed offered plant protection through the insertion of specific ndo genes as visualized by DGGE. The comparison of ndo-DGGE fingerprints of DNA and cDNA revealed that both indigenous and introduced bacteria were able to express the genes coding for PAH-degrading enzymes in the rhizosphere. However, due to the presence of dominant bands in the DNA and cDNA in the inoculated treatments, it can be suggested that specific ndo genes are present which are only expressed under high levels of oil pollution and have an earlier activity, as the band intensity was higher on day 7.
In conclusion, the data reported herein indicate that, due to the previous selection of bacterial consortia able to survive by using petroleum as carbon and nitrogen sources, the addition of petroleum did not change the composition of a bacterial community pre-adapted to PAH, in a greenhouse microcosm experiment over time. However, while the overall bacterial community composition did not significantly shift based on the methods used, changes in the abundance of conjugative plasmids and PAH-degrading genes were observed, indicating that horizontal gene transfer is a crucial parameter in the development of effective PAH-degrading microbial communities occurring in urban mangrove soils. It was shown that mangrove roots provide a suitable environment for the expression of PAH-catabolic genes expression. The results of this study aid in elucidating the response in bacterial community composition to oil contamination in mangrove ecosystems, indicating that a bioaugmentation strategy mediated by highly efficient PDHC can be a useful tool to clean-up the environment, with certain advantages, such as low implementation and maintenance costs. The main novelty of this work regarding the PDHC results is that Comamonas sp. (Betaproteobacteria) and Ochrobactrum sp. (Alphaproteobacteria) carrying IncP-9 plasmids are reported here for the first time. Thus it seems that under condition with strong selective pressure the host range of IncP-9 plasmids is far broader as previously reported.
Concluding remarksDGGE71 is a fingerprinting technique which allows to analyze rapidly and in a cost-effective way the structure and dynamics of microbial communities. Herein DGGE provided important information, which indicated that the abundant bacterial communities analyzed were stable over the time likely due to their pre-adaptation to oil. Commonly to every independent of cultivation technique, DGGE has its limitations such as the limit to detect only the most abundant microbial communities (>1% of the total community), not including some of rare species or those below to the detection limit, which can play an important role in PAH degradation. However, previous work2 based on the study of microbial communities from mangrove contaminated with oil indicated the absence of difference in the results revealed by DGGE and barcoded pyrosequencing. Nevertheless, would be interesting future amplicon analysis by next generation sequencing technologies for comparison with the data shown here and to provide information on the taxonomic affiliation of the dominant bacterial community members.
Based on the knowledge that sometimes a certain gene can be present but not expressed, DGGE analysis of ndo amplified from cDNA, instead only DNA analysis of ndo genes, brought a more complete and useful information, indicating not only the presence of ndo genes but also their expression of SBH has many advantages, as it provides insights into the prevalence and diversity of plasmids and genes in a larger numbers samples. When compared to the conventional PCR, which can also amplify non-specific products, SBH significantly increases the specificity and sensitivity of detection.72 Therefore, herein the PCR-based detection combined with SBH revealed intense shifts in the MGEs of bacterial communities, indicating the increased abundance of MGEs which might be due to the proliferation of bacterial populations carrying MGEs or due to horizontal transfer processes. In summary, this study provided novel data about the host range of IncP-9 catabolic plasmids, which were found for the first time in Comamonas sp. and Ochrobactrum spp. Furthermore, this study demonstrated that the PHDC inoculated in the greenhouse microcosm plants protected the A. schaueriana plants against the phytotoxicity induced by the artificial contamination with petroleum. Our results indicated that studies focusing on PHDC development and its application in bioaugmentation can be an interesting approach to promote mangrove environment detoxification and recovery after an oil spill accident.
Conflicts of interestThe authors declare no conflicts of interest.
Acknowledgments are due to PNPD-CAPES (Brazilian Ministry of Education) and CAPES for scholarship support given to S. Dealtry and A.M. Ghizelini. This study was funded by the Deutsche Forschungsgemeinschaft SM59/4-1 and 4-2 and by FAPERJ-Brazil.