In this study, the performance of the “RESIST-3 O.K.N. K-SeT” (Coris BioConcept, Gembloux, Belgium) immunochromatographic assay was evaluated in 132 Klebsiella pneumoniae comprising 102 carbapenem resistant and 30 carbapenem susceptible isolates. Genotypically known isolates of Gram negative bacteria (n=22) including various species were also tested by the assay as controls. The isolates tested by the immunochromatographic assay and also were run PCR for blaKPC, blaIMP, blaVIM, blaNDM, and blaOXA-48. The rates of blaNDM, blaOXA-48, and blaKPC in carbapenem resistant isolates were found at 52.9%, 39.2%, and 2.0%, respectively. Both blaNDM and blaOXA-48 were found in six (5.9%) isolates. The results of the assay showed 100% concordance with those obtained by PCR in 132K. pneumoniae. The agreement between the two methods was found to be identical at the isolate level. The assay also correctly detected all genotypically known isolates of Escherichia coli, Serratia marcescens, Citrobacter freundii, Enterobacter cloacae, K. pneumoniae carrying blaKPC, blaNDM, and/or blaOXA-48. On the other hand, the assay did not exhibit any cross-reaction in control isolates harboring blaIMP and blaVIM. We conclude that the RESIST-3 O.K.N. K-SeT is a reliable, rapid, and user friendly test and we recommend it for routine diagnostic laboratories.
Multi-drug resistant Klebsiella pneumoniae isolates are of great concern worldwide.1 Although carbapenems are widely used antibiotics in the treatment of infections caused by multi-drug resistant K. pneumoniae, nosocomial infections due to carbapenem resistant K. pneumoniae (CRKp) have also increased dramatically in the last decade.2 The main mechanism of carbapenem resistance in K. pneumoniae has been associated with the production of carbapenem hydrolyzing enzymes. Various types of class A, B, and D type carbapenemases have been identified in K. pneumoniae. KPCs are the most commonly seen class A carbapenemases and endemic in the USA, Colombia, Brasil, Argentina, Italy, Poland, China, Taiwan, Israel and Greece.1–3 Among class B carbapenemases, VIM and IMP have spread in many countries since the early 2000s and are endemic in Southern Europe and Asia-Pacific region.2 Another class B carbapenemase, NDM, has also spread rapidly and become a global threat since 2008.4 In class D, OXA-48 type carbapenemases have become prevalent among K. pneumoniae and other members of Enterobacteriaceae in Turkey, Morocco, Tunisia, Libya, Algeria, Egypt, and India.1–3,28,29
The rapid and accurate laboratory diagnosis of carbapenemase-producing isolates is critical to enable the timely application of infection control measures. Standard susceptibility testing cannot specify the mechanism of carbapenem resistance. An ideal diagnostic method should be rapid and able to detect all types of threatening carbapenemases sensitively and specifically. Currently, there are various approaches to phenotypic tests based on different principles. These include inhibitor based approach (combination disk test, double disk synergy test, gradient diffusion strips), detection based on carbapenem hydrolysis (cloverleaf method, colorimetric assays, Carba NP, Blue-CARBA, carbapenem inhibition method, starch-iodine assay, spectrometry, electrochemical assay).5–10 Although some of these methods exhibit high performance in diagnosis of KPC, IMP, VIM and NDM producers; none of them is highly sensitive and specific for detecting OXA-48 producers.5–11 This may lead to challenges in the diagnosis of OXA-48 producers especially in regions where this type of carbapenemases are prevalent. Moreover most of the methods described above are time-consuming. Recent studies have revealed promising results with immunochromatographic assays for easy and rapid detection of specific carbapenamases.6,12–19
In this study we aimed to evaluate the performance of a newly marketed immunochromatographic assay, “RESIST-3 O.K.N. K-SeT” (Coris BioConcept, Gembloux, Belgium) targeting OXA-48 like, KPC, and NDM type carbapenemases in a collection of carbapenem resistant K. pneumoniae.
Materials and methodsStudy isolatesA total of 132 non-duplicated clinical K. pneumoniae isolates collected in two university hospitals between July 2014 and December 2016 were included. 102 out of 132K. pneumoniae were resistant to ertapenem in routine testing by an automated antimicrobial susceptibility test system (VITEK® 2, bioMérieux, Marcy l’Etoile, France). The resting isolates were susceptible to carbapenems in routine testing (Table 1).
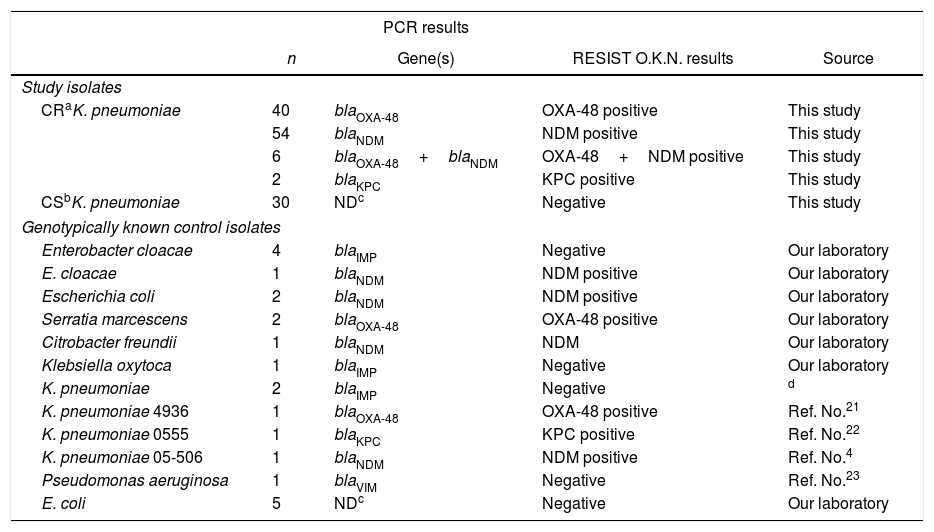
The results of RESIST-3 O.K.N. K-SeT immunochromatographic assay in 132 clinical Klebsiella pneumoniae and 22 control isolates compared to PCR.
| PCR results | ||||
|---|---|---|---|---|
| n | Gene(s) | RESIST O.K.N. results | Source | |
| Study isolates | ||||
| CRaK. pneumoniae | 40 | blaOXA-48 | OXA-48 positive | This study |
| 54 | blaNDM | NDM positive | This study | |
| 6 | blaOXA-48+blaNDM | OXA-48+NDM positive | This study | |
| 2 | blaKPC | KPC positive | This study | |
| CSbK. pneumoniae | 30 | NDc | Negative | This study |
| Genotypically known control isolates | ||||
| Enterobacter cloacae | 4 | blaIMP | Negative | Our laboratory |
| E. cloacae | 1 | blaNDM | NDM positive | Our laboratory |
| Escherichia coli | 2 | blaNDM | NDM positive | Our laboratory |
| Serratia marcescens | 2 | blaOXA-48 | OXA-48 positive | Our laboratory |
| Citrobacter freundii | 1 | blaNDM | NDM | Our laboratory |
| Klebsiella oxytoca | 1 | blaIMP | Negative | Our laboratory |
| K. pneumoniae | 2 | blaIMP | Negative | d |
| K. pneumoniae 4936 | 1 | blaOXA-48 | OXA-48 positive | Ref. No.21 |
| K. pneumoniae 0555 | 1 | blaKPC | KPC positive | Ref. No.22 |
| K. pneumoniae 05-506 | 1 | blaNDM | NDM positive | Ref. No.4 |
| Pseudomonas aeruginosa | 1 | blaVIM | Negative | Ref. No.23 |
| E. coli | 5 | NDc | Negative | Our laboratory |
To assess the specificity of the new immunochromatographic assay, a collection (n=22) of Gram negative bacteria which have been previously characterized by PCR and/or DNA sequencing for the presence of blaKPC, blaIMP, blaVIM, blaNDM, and blaOXA-48 genes were also studied with RESIST-3 O.K.N. K-SeT (Table 1). Escherichia coli ATCC 25922 were used as a reference strain in antimicrobial susceptibility testing.
Bacterial identificationIdentification of the isolates was confirmed by MALDI-TOF mass spectrometry (VITEK® MS, bioMérieux, Marcy l’Etoile, France).
Antimicrobial susceptibility testingStandard disk diffusion test was performed on all isolates to confirm the ertapenem susceptibility results obtained with the VITEK® 2 system.20 In addition, ertapenem MICs were determined by the gradient diffusion strip test (Etest®, bioMérieux, Marcy l’Etoile, France) in the isolates. The European Committee on Antimicrobial Susceptibility Testing (EUCAST v 6.0) standards used as interpretive criteria for antimicrobial susceptibility testing.
Polymerase chain reactionCarbapenemase encoding genes, blaKPC, blaIMP, blaVIM, blaNDM, and blaOXA-48, were investigated by in house single PCR method to confirm phenotypic test results in 132K. pneumoniae isolates.21,24–26
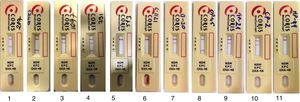
Immunochromatographic assayAll isolates (132K. pneumoniae and 22 genotypically known Gram negative bacteria) were tested by the RESIST-3 O.K.N. K-SeT according to the manufacturer's instructions. Briefly, a single colony on Columbia agar + 5% sheep blood (bioMérieux, Marcy l’Etoile, France) was suspended in 10 drops of lysis buffer. Three drops of the suspension were then added onto the test strip. The results were read with the naked eye within 15minutes at room temperature (Fig. 1).
Evaluation of the test results in the RESIST-3 O.K.N.K-SeT assay. 1; KPC producing K. pneumoniae, 2; OXA-48 producing K. pneumoniae, 3; NDM producing K. pneumoniae, 4; OXA-48 and NDM co-producing K. pneumoniae, 5; Carbapenem susceptible K. pneumoniae as negative control, 6; IMP producing E. cloacae, 7; NDM producing E. cloacae, 8; OXA-48 producing S. marcescens, 9; NDM producing E. coli, 10; NDM producing C. freundii, 11; Carbapenem susceptible E. coli as negative control.
A total of 132K. pneumoniae isolates (102 CRKp and 30 CSKp) were evaluated for the presence of OXA-48 like, KPC, and NDM carbapenemases by the RESIST-3 O.K.N K-SeT immunochtomotographic assay. The results of the RESIST-3 O.K.N. K-SeT assay were compared those obtained by PCR targeting these genes (Table 1). Ertapenem MICs in CRKp isolates ranged from 1 to >32μg/mL (MIC50: 32μg/mL, MIC90: >32μg/mL) by the gradient diffusion strip test. Rates of blaNDM, blaOXA-48, blaKPC, genes in CRKp isolates were found to be 52.9%, 39.2%, and 2.0%, respectively, by PCR. Combined positivity for both blaNDM and blaOXA-48 genes was found in six (5.9%) isolates. The carbapenemase genes blaIMP and blaVIM were not detected in any of 132 study isolates. The results of the RESIST-3 O.K.N. K-SeT assay showed 100% concordance with those obtained by PCR. The agreement between the two methods was found to be identical at the isolate level. The RESIST-3 O.K.N. K-SeT assay did not exhibit any cross-reaction in genotypically known bacteria (n=8) harboring blaIMP and blaVIM genes (Table 1). The results obtained with the assay also showed agreement in genotypically known (n=9) Enterobacteriaceae isolates (Escherichia coli, Serratia marcescens, Citrobacter freundii, Enterobacter cloacae, K. pneumoniae) carrying blaKPC, blaNDM, and/or blaOXA-48 genes (Table 1). False positive result was not observed in five carbapenem susceptible E. coli isolates which were known negative for blaKPC, blaIMP, blaVIM, blaNDM, and blaOXA-48.
DiscussionInfections due to carbapenemase producing K. pneumoniae pose great concerns since they are associated with high morbidity and mortality.1–3 Rapid and specific diagnosis of carbapenemase producers plays a crucial role in preventing the spread of CRKp among hospitalized patients. Although PCR is recommended as ‘gold standard’ in carbapenemase detection,5,18,27 in terms of laboratory use, it has several unfavorable properties that limit its usefulness in many routine diagnostic laboratories.5,27 These properties include difficulty in application, and the need for specific equipment and trained personnel.5,8,9 Moreover, PCR cannot reveal the expression state of a gene. The availability of a reliable, rapid, and a simple phenotypic assay to detect carbapenemases would be of great benefit. Furthermore, such a test should be able to identify most of the prevalent carbapenemases with epidemiological significance. Various phenotypic methods developed so far are considered as reliable assays especially in detecting class A and B carbapenemases by many authors.5–11 However these methods remained unsatisfactory in the diagnosis of OXA-48 producers. A few immunochromatographic assays in the market have been shown as reliable and fast for detecting OXA-48 and/or KPC producers, separately.12–17 Unfortunately, these tests are not able to identify the most common carbapenemases, simultaneously. In this study, we evaluated the performance of a recently introduced RESIST-3 O.K.N. K-SeT immunochromatographic assay. It is the first assay in the market targeting the detection of KPC, NDM, and OXA-48 together.
In our CRKp collection, RESIST-3 O.K.N. K-SeT results exhibited excellent concordance with PCR, yielding 100% sensitivity for the tested isolates (Table 1). The assay also detected five NDM, three OXA-48 and one KPC producers correctly in genotypically known collection of E. cloacae, E. coli, S. marcescens, C. freundii, K. pneumoniae (Table 1). Considering the specificity of the test, all carbapenem susceptible isolates of K. pneumoniae (n=30) and E. coli (n=5) and the isolates harboring blaIMP, blaVIM genes were found to be negative by the assay. These results indicate the high specificity (100%) of RESIST-3 O.K.N. K-SeT. The assay also detected coproduction of NDM and OXA-48 in clinical isolates harboring blaNDM and blaOXA-48 genes. Recent studies have remarked the emergence of K. pneumoniae and other Enterobacteriaceae producing both carbapenemases from Turkey and some other countries.31–40 For that, our result was found to be significant in terms of the reliability of RESIST-3 O.K.N. K-SeT in detecting NDM and OXA-48 co-producers. We have a limited number of KPC producers in our collection. Therefore, it is not easy to reach a conclusion about the reliability of the RESIST-3 O.K.N. K-SeT test in KPC detection. As stated in many reports KPC is not an endemic carbapenemase in our country.1–3,28–30 Two recently published reports have shown that the RESIST-3 O.K.N. K-SeT exhibited the high performance in the detection of KPC producers as well as the producers of NDM and OXA-48.18,19 As concluded by the authors of these two studies we also suggest that the assay has a limitation in terms of detecting other carbapenemases such as IMP and VIM producers. We suggest that there is still a need for different assays in detection the isolates producing carbapenemases other than KPC, NDM, and OXA-48.
Besides the evaluation of RESIST-3 O.K.N. K-SeT performance, our work has also revealed some outputs regarding the prevalent carbapenemases in our CRKp collection. Accordingly, the most common carbapenemase has been NDM (52.9%), followed by OXA-48 (39.2%). These two carbapenemases have been found together in 5.9% of CRKp isolates. We consider these results to be very remarkable in terms of revealing the predominance of NDM in our CRKp collection. Almost all studies published from Turkey so far, have emphasized OXA-48 prevalence in the country.21,28,31,33 Although the presence of various reports from Turkey notifying the emergence of NDM in the country, this is the first report highlighting the very high rate of NDM in an extended collection of CRKp in our region.31–35 Moreover the detection of CRKp isolates harboring NDM together with OXA-48 was considered as a premonitory outcome of the study because it shows the spread of CRKp NDM and OXA-48 co-producers.31–40 On the other hand, this report has been the third one from Turkey presenting the isolation of KPC producing K. pneumoniae, so far.41,42 Kuskucu et al. also isolated KPC enzymes in two E. coli isolates in Turkey.43 In many other countries, including Turkey's neighboring country Greece, the predominant enzyme type is KPC. The absence of any regional spread or institutional outbreaks due to KPC in Turkey may be associated with the already high burden of other resistance determinants among K. pneumoniae isolates, and the genetic failure of the sporadic KPC cases to transfer their plasmids carrying the blaKPC gene.
Finally, we conclude that the RESIST-3 O.K.N.K-SeT assay is a reliable, rapid, user-friendly test and we recommend it for routine diagnostic laboratories especially in regions where OXA-48 and NDM producers are prevalent.
Conflicts of interestThe authors declare that there are no conflicts of interest.
We thank Cigdem Kayacan for granting the OXA-48, IMP-1 and VIM-4 positive strains.
This study was supported by internal funding and partly supported by in-kind grant from Zemed Medikal Tekstil ve Dıs Tic. Ltd. Sti., Istanbul, Turkey.
This study was approved by the Marmara University Clinical Research Ethics Committee (Decision No: 09.2017.224).